8AI5
 
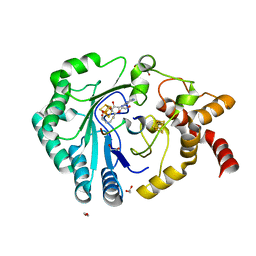 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 6 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
4RP2
 
 | |
4ROY
 
 | |
4ROG
 
 | |
4ROK
 
 | |
4RON
 
 | |
4RVH
 
 | | Crystal structure of MtmC in complex with SAH and TDP-4-keto-D-olivose | | Descriptor: | D-mycarose 3-C-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RP0
 
 | |
4ROO
 
 | |
4ROZ
 
 | |
4RP1
 
 | |
4RO7
 
 | |
4RO8
 
 | |
8CK1
 
 | | Carin 1 bacteriophage tail, connector and tail fibers assembly | | Descriptor: | Connector Protein, Tail Nozzle, Tail fibers Dpo36 | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
8CJZ
 
 | | Carin1 bacteriophage mature capsid | | Descriptor: | Capsid Decoration Protein, Major Capsid Protein, Spike Base Protein | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
2QP6
 
 | |
8BI8
 
 | | Structure of a cyclic beta-hairpin peptide derived from neuronal nitric oxide synthase | | Descriptor: | Nitric oxide synthase, brain | | Authors: | Balboa, J.R, Ostergaard, S, Stromgaard, K, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of a Potent Cyclic Peptide Inhibitor of the nNOS/PSD-95 Interaction.
J.Med.Chem., 66, 2023
|
|
5KWX
 
 | |
8PPG
 
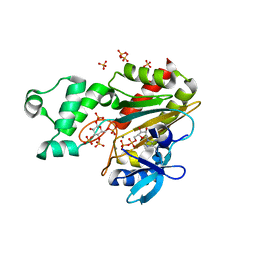 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with beta-D-glucopyranosyl 1,3,4,6-tetrakisphosphate/ADP/Mn after reaction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPJ
 
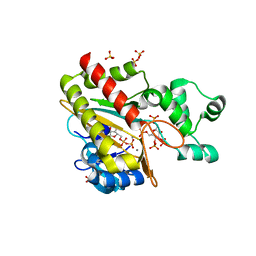 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with beta-D-glucopyranosylmethanol 3,4,6,1'-tetrakisphosphate/ADP/Mn after reaction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
4XPP
 
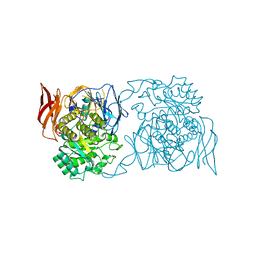 | | Crystal structure of Pedobacter saltans GH31 alpha-galactosidase complexed with D-galactose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase, beta-D-galactopyranose | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
5KX2
 
 | |
5KVN
 
 | |
4U6G
 
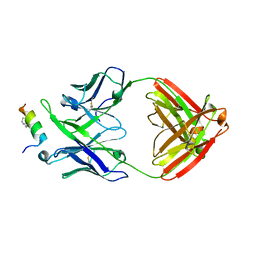 | | Crystal structure of 10E8 Fab in complex with a hydrocarbon-stapled HIV-1 gp41 MPER peptide | | Descriptor: | 10E8 Fab Heavy Chain, 10E8 Fab Light Chain, SAH-MPER(662-683KKK)(B,q) | | Authors: | Ofek, G, Bird, G.H, Walensky, L.D, Kwong, P.D. | | Deposit date: | 2014-07-28 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.2012 Å) | | Cite: | Structural Fidelity and Neutralizing Antibody Binding Capacity of
Hydrocarbon-Stapled HIV-1 Antigens
To be published
|
|
2QOA
 
 | |
