8GXS
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in H-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
6JGR
 
 | | Crystal structure of barley exohydrolaseI W434Y in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6J0E
 
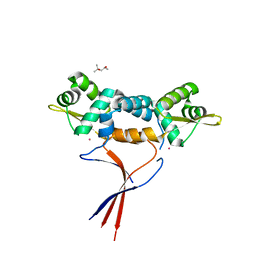 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ARSENIC, Arsenic responsive repressor ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
7DOZ
 
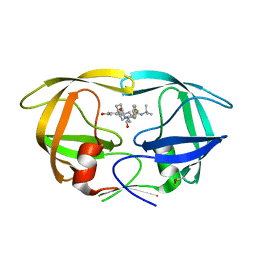 | | HIV-1 Protease D30N mutant in complex with Nelfinavir | | Descriptor: | 2-[2-HYDROXY-3-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-4-PHENYL SULFANYL-BUTYL]-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID TERT-BUTYLAMIDE, Protease | | Authors: | Bihani, S.C, Hosur, M.V. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular basis for reduced cleavage activity and drug resistance in D30N HIV-1 protease.
J.Biomol.Struct.Dyn., 2021
|
|
8GZ3
 
 | | Structure of human phagocyte NADPH oxidase in the resting state | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7D5 Fab heavy chain, ... | | Authors: | Chen, L, Liu, R. | | Deposit date: | 2022-09-24 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human phagocyte NADPH oxidase in the resting state.
Elife, 11, 2022
|
|
6J3Z
 
 | | Structure of C2S1M1-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
3L94
 
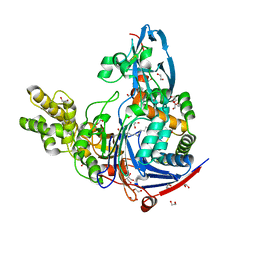 | | Structure of PvdQ covalently acylated with myristate | | Descriptor: | 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase pvdQ subunit alpha, Acyl-homoserine lactone acylase pvdQ subunit beta, ... | | Authors: | Drake, E.J, Gulick, A.M. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization and high-throughput screening of inhibitors of PvdQ, an NTN hydrolase involved in pyoverdine synthesis.
Acs Chem.Biol., 6, 2011
|
|
6J3Y
 
 | | Structure of C2S2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
8USP
 
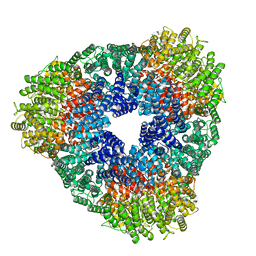 | |
8USQ
 
 | |
6JG6
 
 | | Crystal structure of barley exohydrolaseI W286A mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGT
 
 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with methyl 6-thio-beta-gentiobioside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JHF
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JG1
 
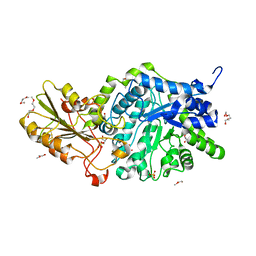 | | Crystal structure of barley exohydrolaseI wildtype in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Barley exohydrolase I, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGG
 
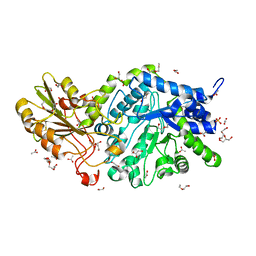 | | Crystal structure of barley exohydrolaseI W434F mutant in complex with methyl 2-thio-beta-sophoroside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JEN
 
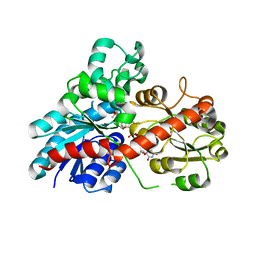 | | Structure of Phytolacca americana UGT2 complexed with UDP-2fluoro-glucose and pterostilbene | | Descriptor: | Glycosyltransferase, Pterostilbene, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2019-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An Ambidextrous Polyphenol GlycosyltransferasePaGT2 fromPhytolacca americana.
Biochemistry, 59, 2020
|
|
8HAP
 
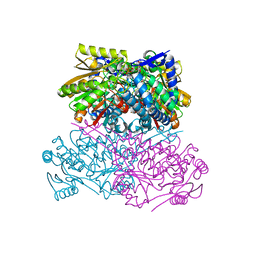 | | Crystal structure of thermostable acetaldehyde dehydrogenase from hyperthermophilic archaeon Sulfolobus tokodaii | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Aldehyde dehydrogenase, SODIUM ION, ... | | Authors: | Mine, S, Nakabayashi, M, Ishikawa, K. | | Deposit date: | 2022-10-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermostable acetaldehyde dehydrogenase from the hyperthermophilic archaeon Sulfolobus tokodaii.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
3LU8
 
 | | Human serum albumin in complex with compound 3 | | Descriptor: | N-[5-(5-{[(2,4-dimethyl-1,3-thiazol-5-yl)sulfonyl]amino}-6-fluoropyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]acetamide, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
6JGK
 
 | | Crystal structure of barley exohydrolaseI W434F mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JFJ
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltohexaose and alpha-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8H0I
 
 | | Cryo-EM structure of APOBEC3G-Vif complex | | Descriptor: | APOBEC3G, CHLORIDE ION, Core binding factor beta, ... | | Authors: | Kouno, T, Shibata, S, Hyun, J, Kim, T.G, Wolf, M. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into RNA bridging between HIV-1 Vif and antiviral factor APOBEC3G.
Nat Commun, 14, 2023
|
|
6JG2
 
 | | Crystal structure of barley exohydrolaseI wildtype in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-nitrophenoxy)-3,5-bis(oxidanyl)oxan-4-yl]sulfanyl-oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Barley exohydrolase I, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGP
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with methyl 6-thio-beta-gentiobioside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGA
 
 | | Crystal structure of barley exohydrolaseI W286F in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-4-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-oxane-2,3,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGL
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with methyl 2-thio-beta-sophoroside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
