1UXC
 
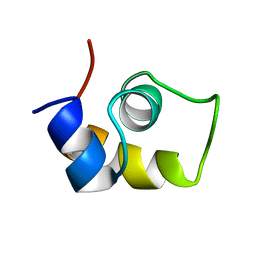 | | FRUCTOSE REPRESSOR DNA-BINDING DOMAIN, NMR, MINIMIZED STRUCTURE | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
1FJ0
 
 | |
1IXM
 
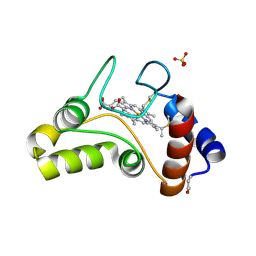
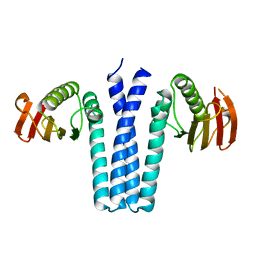 | | CRYSTAL STRUCTURE OF SPOOB FROM BACILLUS SUBTILIS | | Descriptor: | PROTEIN (SPORULATION RESPONSE REGULATORY PROTEIN) | | Authors: | Varughese, K.I. | | Deposit date: | 1998-11-06 | | Release date: | 1998-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase.
Mol.Cell, 2, 1998
|
|
2ZNP
 
 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist TIPP204 | | Descriptor: | (2S)-2-{4-butoxy-3-[({[2-fluoro-4-(trifluoromethyl)phenyl]carbonyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Oyama, T, Hirakawa, Y, Nagasawa, N, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1P7E
 
 | |
1JCJ
 
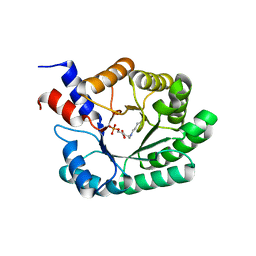 | | OBSERVATION OF COVALENT INTERMEDIATES IN AN ENZYME MECHANISM AT ATOMIC RESOLUTION | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, DEOXYRIBOSE-PHOSPHATE ALDOLASE | | Authors: | Heine, A, DeSantis, G, Luz, J.G, Mitchell, M, Wong, C.-H, Wilson, I.A. | | Deposit date: | 2001-06-09 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
Science, 294, 2001
|
|
1RMU
 
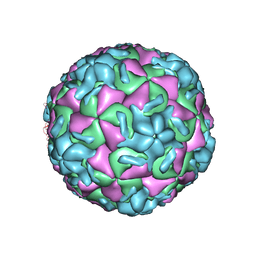 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
3DDX
 
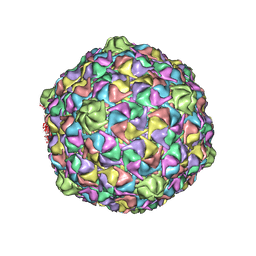 | | HK97 bacteriophage capsid Expansion Intermediate-II model | | Descriptor: | Major capsid protein | | Authors: | Lee, K.K, Gan, L, Conway, J.F, Hendrix, R.W, Steven, A.C, Johnson, J.E. | | Deposit date: | 2008-06-06 | | Release date: | 2008-11-04 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Virus capsid expansion driven by the capture of mobile surface loops.
Structure, 16, 2008
|
|
3NFE
 
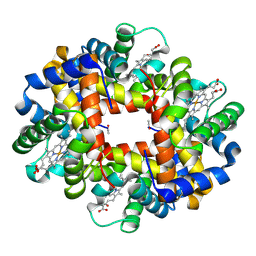 | | The crystal structure of hemoglobin I from trematomus newnesi in deoxygenated state | | Descriptor: | Hemoglobin subunit alpha-1, Hemoglobin subunit beta-1/2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vergara, A, Vitagliano, L, Merlino, A, Sica, F, Marino, K, Mazzarella, L. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | An order-disorder transition plays a role in switching off the root effect in fish hemoglobins.
J.Biol.Chem., 285, 2010
|
|
3E3U
 
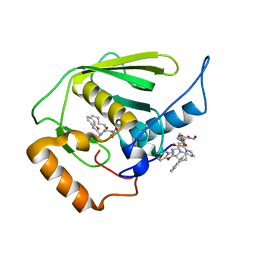 | | Crystal structure of Mycobacterium tuberculosis peptide deformylase in complex with inhibitor | | Descriptor: | N-[(2R)-2-{[(2S)-2-(1,3-benzoxazol-2-yl)pyrrolidin-1-yl]carbonyl}hexyl]-N-hydroxyformamide, NICKEL (II) ION, Peptide deformylase | | Authors: | Meng, W, Xu, M, Pan, S, Koehn, J. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1FCC
 
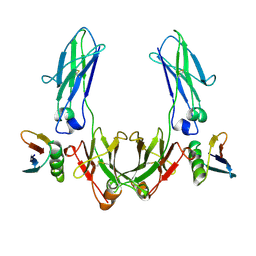 | | CRYSTAL STRUCTURE OF THE C2 FRAGMENT OF STREPTOCOCCAL PROTEIN G IN COMPLEX WITH THE FC DOMAIN OF HUMAN IGG | | Descriptor: | IGG1 MO61 FC, STREPTOCOCCAL PROTEIN G (C2 FRAGMENT) | | Authors: | Sauer-Eriksson, A.E, Kleywegt, G.J, Uhlen, M, Jones, T.A. | | Deposit date: | 1995-01-17 | | Release date: | 1995-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG.
Structure, 3, 1995
|
|
1IXO
 
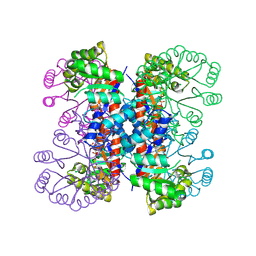 | | Enzyme-analogue substrate complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | Pyridoxine 5'-Phosphate synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
1SRR
 
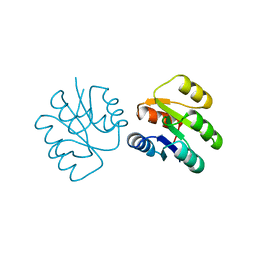 | | CRYSTAL STRUCTURE OF A PHOSPHATASE RESISTANT MUTANT OF SPORULATION RESPONSE REGULATOR SPO0F FROM BACILLUS SUBTILIS | | Descriptor: | CALCIUM ION, SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Whiteley, J.M, Hoch, J.A, Zapf, J, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1996-04-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis.
Structure, 4, 1996
|
|
1DE4
 
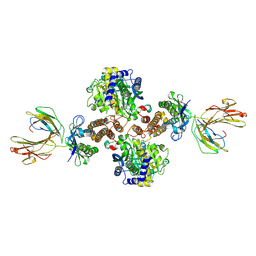 | | HEMOCHROMATOSIS PROTEIN HFE COMPLEXED WITH TRANSFERRIN RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-2-MICROGLOBULIN, CALCIUM ION, ... | | Authors: | Bennett, M.J, Lebron, J.A, Bjorkman, P.J. | | Deposit date: | 1999-11-12 | | Release date: | 2000-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor.
Nature, 403, 2000
|
|
1G74
 
 | |
1G7N
 
 | |
1LT6
 
 | |
3NOU
 
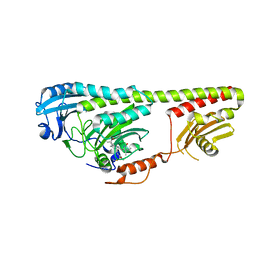 | |
2KZG
 
 | | A Transient and Low Populated Protein Folding Intermediate at Atomic Resolution | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Religa, T.L, Banachewicz, W, Fersht, A.R, Kay, L.E. | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A transient and low-populated protein-folding intermediate at atomic resolution.
Science, 329, 2010
|
|
3NOT
 
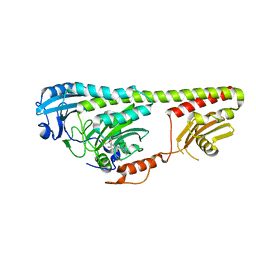 | |
1UZM
 
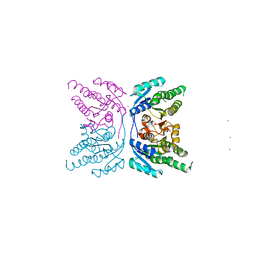 | | MabA from Mycobacterium tuberculosis | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER PROTEIN] REDUCTASE, CESIUM ION | | Authors: | Cohen-Gonsaud, M, Ducasse, S, Quemard, A, Labesse, G. | | Deposit date: | 2004-03-14 | | Release date: | 2005-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of Maba from Mycobacterium Tuberculosis, a Reductase Involved in Long-Chain Fatty Acid Biosynthesis.
J.Mol.Biol., 320, 2002
|
|
2AKA
 
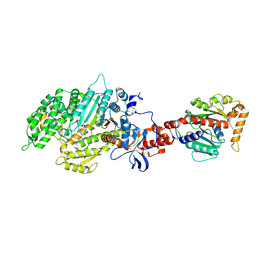 | | Structure of the nucleotide-free myosin II motor domain from Dictyostelium discoideum fused to the GTPase domain of dynamin 1 from Rattus norvegicus | | Descriptor: | Dynamin-1, LINKER, myosin II heavy chain | | Authors: | Reubold, T.F, Eschenburg, S, Becker, A, Leonard, M, Schmid, S.L, Vallee, R.B, Kull, F.J, Manstein, D.J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GTPase domain of rat dynamin 1.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AVF
 
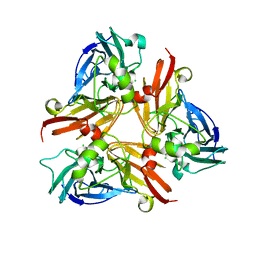 | | Crystal Structure of C-terminal Desundecapeptide Nitrite Reductase from Achromobacter cycloclastes | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Li, H.T, Chang, T, Chang, W.C, Chen, C.J, Liu, M.Y, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2005-08-30 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of C-terminal desundecapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 338, 2005
|
|
2AYY
 
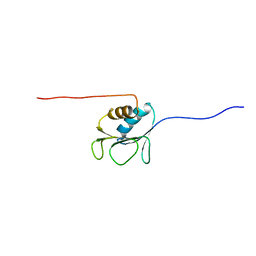 | | Solution structure of the E.coli RcsC C-terminus (residues 700-816) containing linker region | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AYZ
 
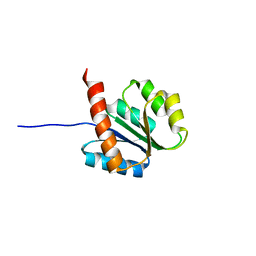 | | Solution structure of the E.coli RcsC C-terminus (residues 817-949) containing phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
