9CV0
 
 | | Bufavirus 1 at pH 7.4 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-27 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
7B87
 
 | | Notum-Fragment074 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
8WDN
 
 | | Crystal structure of PDE4D complexed with 7b-1 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-7-ethoxy-2-(3-ethoxy-4-methoxy-phenyl)-3-(hydroxymethyl)-2,3-dihydro-1-benzofuran-5-yl]propan-1-ol, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of Dihydrobenzofuran Neolignans as Novel PDE4 Inhibitors and Evaluation of Antiatopic Dermatitis Efficacy in DNCB-Induced Mice Model.
J.Med.Chem., 67, 2024
|
|
8R2F
 
 | | human carbonic anhydrase II in complex with 4-(((dimethoxyphosphoryl)(4-((dimethoxyphosphoryl)((4-sulfamoylphenyl)amino)methyl)phenyl)methyl)amino)phenyl sulfuramidite | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(~{S})-dimethoxyphosphoryl-[4-[(~{S})-dimethoxyphosphoryl-[(4-sulfamoylphenyl)amino]methyl]phenyl]methyl]amino]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-11-04 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Sulfonamide-incorporated bis( alpha-aminophosphonates) as promising carbonic anhydrase inhibitors: Design, synthesis, biological evaluation, and X-ray crystallographic studies.
Arch Pharm, 357, 2024
|
|
6X4P
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 28) | | Descriptor: | (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
7P2C
 
 | | F(M197)H mutant structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV by fixed-target serial synchrotron crystallography (room temperature, 26keV) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Guenther, S, Meents, A, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2021-07-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | X-ray structure of the Rhodobacter sphaeroides reaction center with an M197 Phe→His substitution clarifies the properties of the mutant complex.
Iucrj, 9, 2022
|
|
8U27
 
 | | Bcl-2-xL complexed with compound 35 | | Descriptor: | Apoptosis regulator Bcl-2, Bcl-2-like protein 1 chimera, propan-2-yl {4-[(5S)-1-(4-bromobenzoyl)-5-phenyl-4,5-dihydro-1H-pyrazol-3-yl]phenyl}carbamate | | Authors: | Rizo, J, Pan, Y.-Z. | | Deposit date: | 2023-09-05 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights for selective disruption of Beclin 1 binding to Bcl-2.
Commun Biol, 6, 2023
|
|
6AY0
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) soaked with peptide | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
8EQG
 
 | |
8RBE
 
 | | Crystal structure of Mycobacterium tuberculosis MmaA1 in apo form | | Descriptor: | GLYCEROL, Mycolic acid methyltransferase MmaA1 | | Authors: | Kobakhidze, G, Wachelder, L, Chaudhary, B, Mazumdar, P.A, Madhurantakam, C, Dong, G. | | Deposit date: | 2023-12-04 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the mycolic acid methyl transferase 1 (MmaA1) from Mycobacterium tuberculosis in the apo-form and in complex with different cofactors reveal unique features for substrate binding.
J.Biomol.Struct.Dyn., 2025
|
|
6OGL
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P51) in complex with GRL-003 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
8EO4
 
 | |
8EQK
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAACCGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-10-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8WAI
 
 | | Structure of RvY_06210 at 1.45 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, RvY_06210, ZINC ION | | Authors: | Kato, S, Fukuda, Y, Inoue, T. | | Deposit date: | 2023-09-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Metabolite phosphatase from anhydrobiotic tardigrades.
Febs J., 291, 2024
|
|
8EQL
 
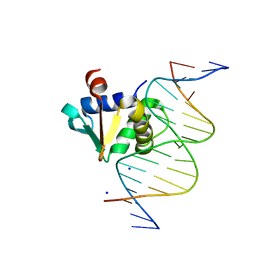 | |
6QVD
 
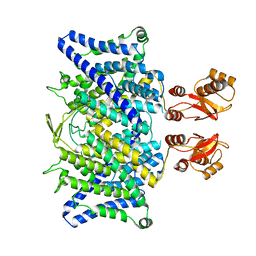 | |
7MOL
 
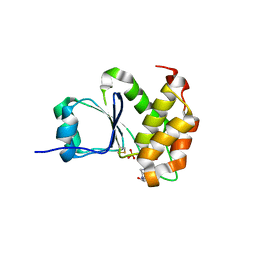 | |
6QOT
 
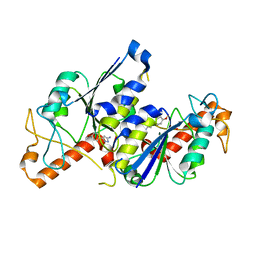 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 23 (3-Amino-5-(4-methoxyphenyl)pyrazole) | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-methoxyphenyl)-1~{H}-pyrazol-5-amine, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
7RR3
 
 | | Structure of Deep-Sea Phage NrS-1 Primase-Polymerase N300 in complex with calcium and ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, CALCIUM ION, Primase | | Authors: | Wang, L, Yu, C, Sliz, P. | | Deposit date: | 2021-08-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Molecular Dissection of the Primase and Polymerase Activities of Deep-Sea Phage NrS-1 Primase-Polymerase.
Front Microbiol, 12, 2021
|
|
8OV4
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Toyocamycin | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8OTO
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with AMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8OV1
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
6WRN
 
 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) C70A variant bound to 3-nitro-tyrosine | | Descriptor: | META-NITRO-TYROSINE, SODIUM ION, Tyrosine--tRNA ligase | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
8OT0
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with MTA and glycine | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Falke, S. | | Deposit date: | 2023-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
6X4Q
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 33) | | Descriptor: | (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
