7P00
 
 | | Human Neurokinin 1 receptor (NK1R) substance P Gq chimera (mGsqi) complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Thom, C, Ehrenmann, J, Vacca, S, Waltenspuhl, Y, Schoppe, J, Medalia, O, Pluckthun, A. | | Deposit date: | 2021-06-29 | | Release date: | 2021-12-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structures of neurokinin 1 receptor in complex with G q and G s proteins reveal substance P binding mode and unique activation features.
Sci Adv, 7, 2021
|
|
7P02
 
 | | Human Neurokinin 1 receptor (NK1R) substance P Gs complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Thom, C, Ehrenmann, J, Vacca, S, Waltenspuhl, Y, Schoppe, J, Medalia, O, Pluckthun, A. | | Deposit date: | 2021-06-29 | | Release date: | 2021-12-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structures of neurokinin 1 receptor in complex with G q and G s proteins reveal substance P binding mode and unique activation features.
Sci Adv, 7, 2021
|
|
4X2I
 
 | | Discovery of benzotriazolo diazepines as orally-active inhibitors of BET bromodomains: Crystal structure of BRD4 with CPI-13 | | Descriptor: | (4R)-6-(4-chlorophenyl)-1,4-dimethyl-5,6-dihydro-4H-[1,2,4]triazolo[4,3-a][1,5]benzodiazepine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Bellon, S.F, Jayaram, H, Poy, F. | | Deposit date: | 2014-11-26 | | Release date: | 2015-11-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of Benzotriazolo[4,3-d][1,4]diazepines as Orally Active Inhibitors of BET Bromodomains.
Acs Med.Chem.Lett., 7, 2016
|
|
8Z7N
 
 | | Structure of HIV-1 CH119 SOSIP.664 trimer in complex with CD4 molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, T-cell surface glycoprotein CD4 | | Authors: | Li, D, Wang, T. | | Deposit date: | 2024-04-20 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Intermediate open state of CD4-bound HIV-1 env heterotrimers in asia CRFs.
Biochem.Biophys.Res.Commun., 725, 2024
|
|
8F00
 
 | | Lysozyme Anomalous Dataset at 293 K and 12 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6YYI
 
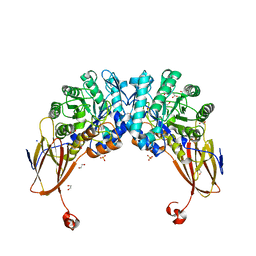 | | Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase, CITRIC ACID, ... | | Authors: | Lafite, P, Daniellou, R, Bretagne, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of Dictyoglomus thermophilum beta-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis.
Biochimie, 181, 2020
|
|
8ZI0
 
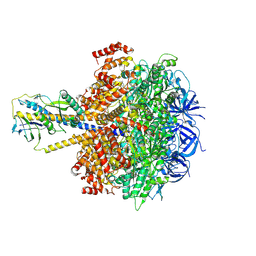 | | Cryo-EM reveals transition states of the Acinetobacter baumannii F1-ATPase rotary subunits gamma and epsilon and novel compound targets - Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Le, K.C.M, Wong, C.F, Gruber, G. | | Deposit date: | 2024-05-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Cryo-EM reveals transition states of the Acinetobacter baumannii F 1 -ATPase rotary subunits gamma and epsilon , unveiling novel compound targets.
Faseb J., 38, 2024
|
|
6Z9I
 
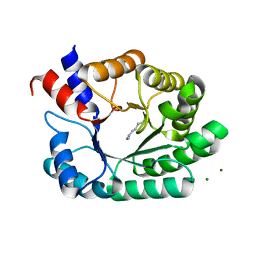 | | Escherichia coli D-2-deoxyribose-5-phosphate aldolase - N21K mutant complex with reaction products | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribose-phosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-18 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Substrate specificity of 2-deoxy-D-ribose 5-phosphate aldolase (DERA) assessed by different protein engineering and machine learning methods.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
8F2Q
 
 | |
6QSI
 
 | |
8Z43
 
 | | Beta-galactosidase from Bacteroides xylanisolvens (E350G, ligand-free) | | Descriptor: | Beta-galactosidase, GLYCEROL | | Authors: | Nakajima, M, Motouchi, S, Kobayashi, K. | | Deposit date: | 2024-04-16 | | Release date: | 2025-01-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure and function of a beta-1,2-galactosidase from Bacteroides xylanisolvens, an intestinal bacterium.
Commun Biol, 8, 2025
|
|
6OGQ
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-003 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
7P7N
 
 | |
7P7O
 
 | | X-RAY CRYSTAL STRUCTURE OF SPOROSARCINA PASTEURII UREASE INHIBITED BY THE GOLD(I)-DIPHOSPHINE COMPOUND Au(PEt3)2Cl DETERMINED AT 1.87 ANGSTROMS | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Ciurli, S, Cianci, M, Messori, L, Massai, L. | | Deposit date: | 2021-07-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Medicinal Au(I) compounds targeting urease as prospective antimicrobial agents: unveiling the structural basis for enzyme inhibition.
Dalton Trans, 50, 2021
|
|
6YYH
 
 | | Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum in ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase, CITRIC ACID, ... | | Authors: | Lafite, P, Daniellou, R, Bretagne, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of Dictyoglomus thermophilum beta-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis.
Biochimie, 181, 2020
|
|
6QOO
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 18 (Isoxazole-5-carbothioamide) | | Descriptor: | 1,2-oxazole-5-carbothioamide, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6QOU
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 24 (Indole-6-boronic acid) | | Descriptor: | 1~{H}-indol-6-ylboronic acid, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6QTY
 
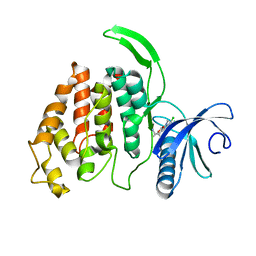 | |
8KB7
 
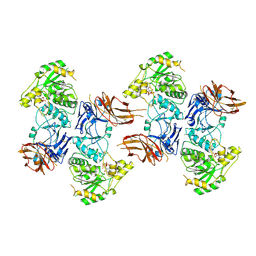 | | Crystal structure of UDP/mannose-bound AGO61/beta-1,4-N-Acetylglucosaminyltransferase 2 (POMGNT2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2, ... | | Authors: | Satoh, T, Umezawa, F, Yagi, H, Kato, K. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of UDP/mannose-bound AGO61/beta-1,4-N-Acetylglucosaminyltransferase 2 (POMGNT2)
To Be Published
|
|
8F1D
 
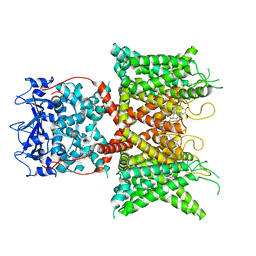 | | Voltage-gated potassium channel Kv3.1 apo | | Descriptor: | CHOLESTEROL, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1, ... | | Authors: | Chen, Y, Ishchenko, A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Identification and structural and biophysical characterization of a positive modulator of human Kv3.1 channels.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6QOA
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 5 (Isoxazole-5-carboxamide) | | Descriptor: | 1,2-oxazole-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6QOF
 
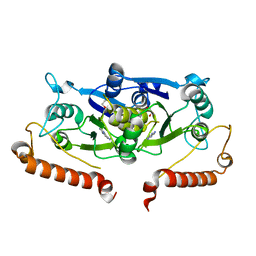 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 9 (Adenine) | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6OGS
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-001 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6QOC
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 6 (2-(1,3-oxazol-5-yl) aniline) | | Descriptor: | 2-(1,3-oxazol-5-yl)aniline, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
5XF2
 
 | | Crystal structure of SeMet-HldC from Burkholderia pseudomallei | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-04-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Expression and crystallographic studies of D-glycero-beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
