2X7F
 
 | | Crystal structure of the kinase domain of human Traf2- and Nck- interacting Kinase with Wee1Chk1 inhibitor | | Descriptor: | 9-HYDROXY-4-PHENYLPYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, SODIUM ION, TRAF2 AND NCK-INTERACTING PROTEIN KINASE | | Authors: | Vollmar, M, Alfano, I, Shrestha, B, Bray, J, Muniz, J.R.C, Roos, A, Filippakopoulos, P, Burgess-Brown, N, Ugochukwu, E, Gileadi, O, Phillips, C, Mahajan, P, Pike, A.C.W, Fedorov, O, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Kinase Domain of Human Traf2- and Nck-Interacting Kinase with Wee1Chk1 Inhibitor
To be Published
|
|
2A26
 
 | | Crystal structure of the N-terminal, dimerization domain of Siah Interacting Protein | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Calcyclin-binding protein, SULFATE ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
5ZPW
 
 | | Generation of a long-acting fusion inhibitor against HIV-1 | | Descriptor: | MET-THR-TRP-GLU-GLU-TRP-ASP-MK8-LYS-ILE-GLU-MK8-TYR-THR-MK8-LYS-ILE-GLU-MK8-LEU-ILE-LYS-LYS-SER, Transmembrane protein gp41 | | Authors: | Guo, Y, Shi, X.L. | | Deposit date: | 2018-04-16 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Generation of a long-acting fusion inhibitor against HIV-1.
Medchemcomm, 9, 2018
|
|
2DNY
 
 | | Solution structure of the third RNA binding domain of FBP-interacting repressor, SIAHBP1 | | Descriptor: | Fuse-binding protein-interacting repressor, isoform b | | Authors: | Suzuki, S, Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-27 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third RNA binding domain of FBP-interacting repressor, SIAHBP1
To be Published
|
|
1LXD
 
 | | CRYSTAL STRUCTURE OF THE RAS INTERACTING DOMAIN OF RALGDS, A GUANINE NUCLEOTIDE DISSOCIATION STIMULATOR OF RAL PROTEIN | | Descriptor: | RALGDSB | | Authors: | Huang, L, Weng, X.W, Hofer, F, Martin, G.S, Kim, S.H. | | Deposit date: | 1997-03-05 | | Release date: | 1998-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of the Ras-interacting domain of RalGDS.
Nat.Struct.Biol., 4, 1997
|
|
6IW6
 
 | | Crystal structure of the Lin28-interacting module of human TUT4 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Terminal uridylyltransferase 4,Terminal uridylyltransferase 4, ... | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2018-12-04 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of the Lin28-interacting module of human terminal uridylyltransferase that regulates let-7 expression.
Nat Commun, 10, 2019
|
|
2VN8
 
 | | Crystal structure of human Reticulon 4 interacting protein 1 in complex with NADPH | | Descriptor: | CITRIC ACID, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pike, A.C.W, Guo, K, Elkins, J, Ugochukwu, E, Roos, A.K, Filippakopoulos, P, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2008-01-31 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Reticulon 4 Interacting Protein 1 in Complex with Nadph
To be Published
|
|
5WVD
 
 | | Structure of Mnk1 in complex with DS12881479 | | Descriptor: | 1-methyl-N-(5-phenyl-1,3-thiazol-2-yl)piperidine-4-carboxamide, MAP kinase interacting serine/threonine kinase 1, SULFATE ION | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2016-12-24 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel inhibitor stabilizes the inactive conformation of MAPK-interacting kinase 1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7EU4
 
 | |
2A7O
 
 | | Solution Structure of the hSet2/HYPB SRI domain | | Descriptor: | Huntingtin interacting protein B | | Authors: | Li, M, Phatnani, H.P, Guan, Z, Sage, H, Greenleaf, A, Zhou, P. | | Deposit date: | 2005-07-05 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Set2 Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1X61
 
 | |
6XUB
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with monovinyl monopropionyl deuteroheme | | Descriptor: | Chlorite dismutase, harderoheme (III) | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
6XUC
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Chlorite dismutase | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8702 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
6SZJ
 
 | |
6SZE
 
 | |
2L6U
 
 | | Solution NMR Structure of Med25(391-543) Comprising the Activator-Interacting Domain (ACID) of Human Mediator Subuniti 25. Northeast Structural Genomics Consortium Target HR6188A | | Descriptor: | Mediator complex subunit MED25 | | Authors: | Eletsky, A, Ryuechan, W.T, Sukumaran, D.K, Shastry, R, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of MED25(391-543) comprising the activator-interacting domain (ACID) of human mediator subunit 25.
J.Struct.Funct.Genom., 12, 2011
|
|
2Y7X
 
 | | The discovery of potent and long-acting oral factor Xa inhibitors with tetrahydroisoquinoline and benzazepine P4 motifs | | Descriptor: | 6-CHLORO-N-[(3S)-1-(5-FLUORO-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL)-2-OXO-PYRROLIDIN-3-YL]NAPHTHALENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Watson, N.S, Adams, C, Belton, D, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Harling, J.D, Irving, W.R, Irvine, S, Kleanthous, S, McLay, I.M, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Discovery of Potent and Long-Acting Oral Factor Xa Inhibitors with Tetrahydroisoquinoline and Benzazepine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4FXB
 
 | | Crystal structure of CYP105N1 from Streptomyces coelicolor: a cytochrome P450 oxidase in the coelibactin siderophore biosynthetic pathway | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Hong, M.K, Lim, Y.R, Kim, J.K, Kim, D.H, Kang, L.W. | | Deposit date: | 2012-07-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of cytochrome P450 CYP105N1 from Streptomyces coelicolor, an oxidase in the coelibactin siderophore biosynthetic pathway
Arch.Biochem.Biophys., 528, 2012
|
|
7BBO
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
7AV9
 
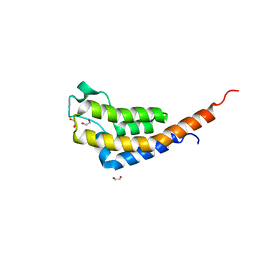 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2
To Be Published
|
|
7BBP
 
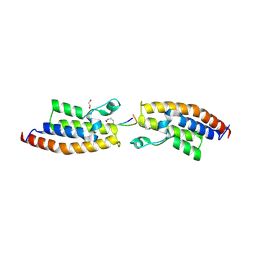 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone H4, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac
To Be Published
|
|
2C4C
 
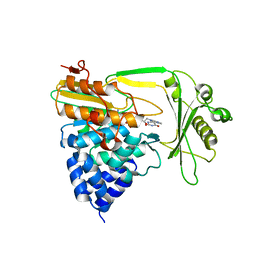 | | Crystal structure of the NADPH-treated monooxygenase domain of MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9-INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-10-18 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2GZD
 
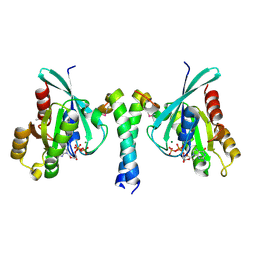 | | Crystal Structure of Rab11 in Complex with Rab11-FIP2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab11 family-interacting protein 2, ... | | Authors: | Khan, A.R. | | Deposit date: | 2006-05-11 | | Release date: | 2006-08-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of rab11 in complex with rab11 family interacting protein 2.
Structure, 14, 2006
|
|
2HW7
 
 | | Crystal Structure of Mnk2-D228G in complex with Staurosporine | | Descriptor: | MAP kinase-interacting serine/threonine-protein kinase 2, STAUROSPORINE, ZINC ION | | Authors: | Jauch, R, Wahl, M.C. | | Deposit date: | 2006-08-01 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mitogen-activated protein kinases interacting kinases are autoinhibited by a reprogrammed activation segment.
Embo J., 25, 2006
|
|
2HW6
 
 | | Crystal structure of Mnk1 catalytic domain | | Descriptor: | MAP kinase-interacting serine/threonine-protein kinase 1, SULFATE ION | | Authors: | Jauch, R, Wahl, M.C. | | Deposit date: | 2006-08-01 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mitogen-activated protein kinases interacting kinases are autoinhibited by a reprogrammed activation segment.
Embo J., 25, 2006
|
|
