4ABJ
 
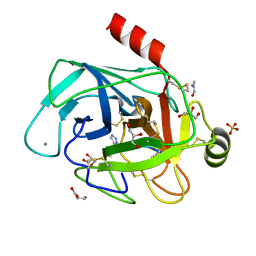 | | Co-complex structure of bovine trypsin with a modified Bowman-Birk inhibitor (IcA)SFTI-1(1,14), that was 1,5-disubstituted with 1,2,3- trizol to mimic a cis amide bond | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, DIMETHYLFORMAMIDE, ... | | Authors: | Schmelz, S, Empting, M, Tischler, M, Nasu, D, Heinz, D, Kolmar, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Braces for the Peptide Backbone: Insights Into Structure-Activity Relation-Ships of Protease Inhibitor Mimics with Locked Amide Conformations
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
5G5D
 
 | | Crystal Structure of the CohScaC2-XDocCipA type II complex from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSOMAL-SCAFFOLDING PROTEIN A, CELLULOSOME ANCHORING PROTEIN COHESIN REGION | | Authors: | Carvalho, A.L, A Bras, J.L, Najmudin, S.H, Pinheiro, B.A, Fontes, C.M.G.A. | | Deposit date: | 2016-05-23 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
1XFO
 
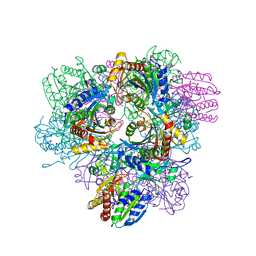 | |
4DNQ
 
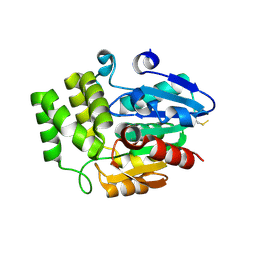 | | Crystal Structure of DAD2 S96A mutant | | Descriptor: | DAD2 | | Authors: | Hamiaux, C. | | Deposit date: | 2012-02-08 | | Release date: | 2012-11-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | DAD2 Is an alpha/beta Hydrolase likely to Be Involved in the Perception of the Plant Branching Hormone, Strigolactone
Curr.Biol., 22, 2012
|
|
8FAV
 
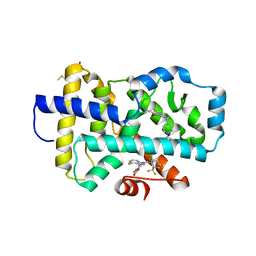 | |
4DNP
 
 | | Crystal Structure of DAD2 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, DAD2, GLYCEROL | | Authors: | Hamiaux, C. | | Deposit date: | 2012-02-08 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DAD2 Is an alpha/beta Hydrolase likely to Be Involved in the Perception of the Plant Branching Hormone, Strigolactone
Curr.Biol., 22, 2012
|
|
2AAX
 
 | | Mineralocorticoid Receptor Double Mutant with Bound Cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, Mineralocorticoid receptor, SULFATE ION | | Authors: | Bledsoe, R.K, Madauss, K.P, Holt, J.A, Apolito, C.J, Lambert, M.H, Pearce, K.H, Stanley, T.B, Stewart, E.L, Trump, R.P, Willson, T.M, Williams, S.P. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Ligand-mediated Hydrogen Bond Network Required for the Activation of the Mineralocorticoid Receptor
J.Biol.Chem., 280, 2005
|
|
3LGU
 
 | | Y162A mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
6TS8
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant G177C/A786C. | | Descriptor: | UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M, Chandran, A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
2NX5
 
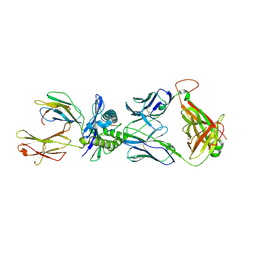 | | Crystal structure of ELS4 TCR bound to HLA-B*3501 presenting EBV peptide EPLPQGQLTAY at 1.7A | | Descriptor: | Beta-2-microglobulin, EBV peptide, EPLPQGQLTAY, ... | | Authors: | Tynan, F.E, Reid, H.H, Rossjohn, J. | | Deposit date: | 2006-11-16 | | Release date: | 2007-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule
Nat.Immunol., 8, 2007
|
|
3LGT
 
 | |
2OL6
 
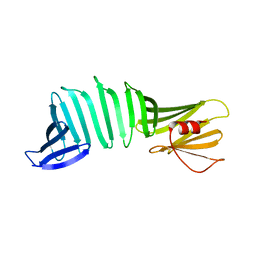 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-01-18 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
2OL8
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-01-18 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
2NW2
 
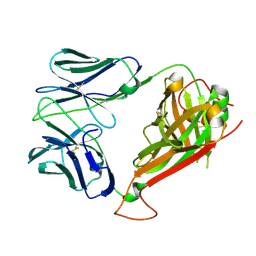 | | Crystal structure of ELS4 TCR at 1.4A | | Descriptor: | ELS4 TCR alpha chain, ELS4 TCR beta chain | | Authors: | Tynan, F.E, Reid, H.H, Rossjohn, J. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule
Nat.Immunol., 8, 2007
|
|
2NW3
 
 | | Crystal structure of HLA-B*3508 presenting EBV peptide EPLPQGQLTAY at 1.7A | | Descriptor: | Beta-2-microglobulin, EBV peptide EPLPQGQLTAY, HLA class I histocompatibility antigen, ... | | Authors: | Tynan, F.E, Reid, H.H, Rossjohn, J. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule
Nat.Immunol., 8, 2007
|
|
2OL7
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-01-18 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
6TBX
 
 | | Trypanosoma brucei PTR1 (TbPTR1) in complex with a tricyclic-based inhibitor | | Descriptor: | 2,4-bis(azanyl)-9~{H}-pyrimido[4,5-b]indol-6-ol, ACETATE ION, GLYCEROL, ... | | Authors: | Landi, G, Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-11-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution crystal structure of Trypanosoma brucei pteridine reductase 1 in complex with an innovative tricyclic-based inhibitor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2OY1
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-02-21 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
4Z6W
 
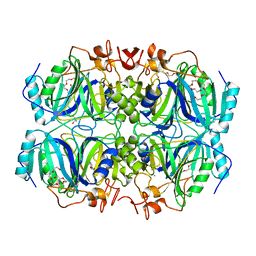 | | Structure of H200N variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-nitrocatechol at 1.57 Ang resolution | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
4Z6P
 
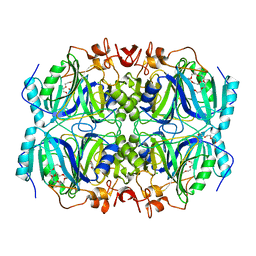 | | Structure of H200Q variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with HPCA at 1.75 Ang resolution | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
4Z6L
 
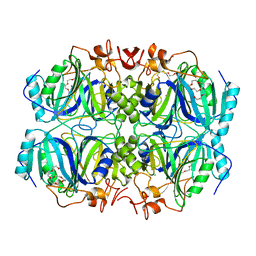 | | Structure of H200E variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum at 1.65 Ang resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
4Z6Z
 
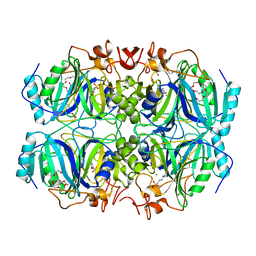 | | Structure of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-sulfonyl catechol at 1.52 Ang resolution | | Descriptor: | 3,4-dihydroxybenzenesulfonic acid, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CALCIUM ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
4Z6S
 
 | | Structure of H200Q variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-sulfonyl catechol at 1.42 Ang resolution | | Descriptor: | 3,4-dihydroxybenzenesulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
4Z6U
 
 | | Structure of H200E variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-nitrocatechol at 1.48 Ang resolution | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
9EOQ
 
 | | Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA | | Descriptor: | DNA (42-MER), DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3') | | Authors: | Ali, K, Georg, K, Volodymyr, M, Johanna, G, Maximilian, N.H, Lukas, K, Simone, C, Hendrik, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM.
Nano Lett., 24, 2024
|
|
