4HNV
 
 | | Crystal structure of R54E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
4HNT
 
 | | crystal structure of F403A mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
5A50
 
 | |
4XCE
 
 | |
4XCC
 
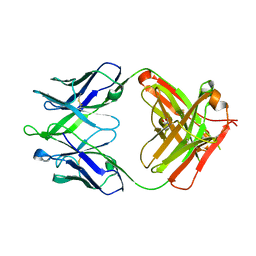 | |
9F5Y
 
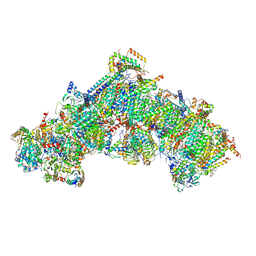 | | Structure of the Chlamydomonas reinhardtii respiratory complex I from respiratory supercomplex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Waltz, F, Righetto, R, Kotecha, A, Engel, B.D. | | Deposit date: | 2024-04-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | In-cell architecture of the mitochondrial respiratory chain.
Science, 387, 2025
|
|
6CUG
 
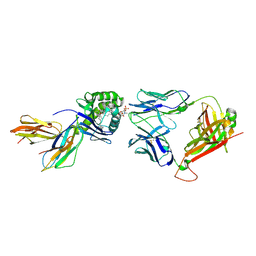 | | Crystal structure of BC8B TCR-CD1b-PC complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Shahine, A.E, Rossjohn, J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids.
Nat Commun, 10, 2019
|
|
4R6W
 
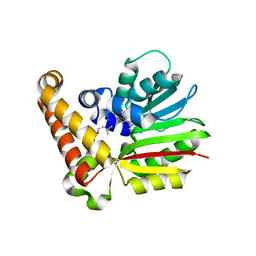 | |
9F5X
 
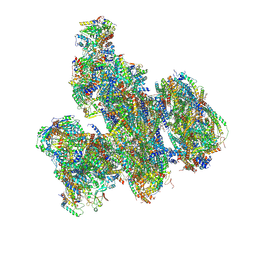 | | Structure of the Chlamydomonas reinhardtii respiratory supercomplex I1 III2 IV2 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Waltz, F, Righetto, R, Kotecha, A, Engel, B.D. | | Deposit date: | 2024-04-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | In-cell architecture of the mitochondrial respiratory chain.
Science, 387, 2025
|
|
8WCR
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
5WP4
 
 | |
9BR4
 
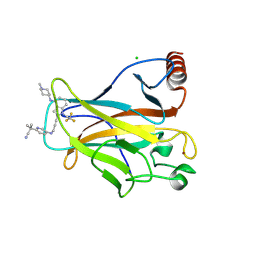 | | Crystal structure of p53 Y220C mutant in complex with PC-9859 | | Descriptor: | 2-methyl-2-{5-[(3-{4-[(1-methylpiperidin-4-yl)amino]-1-(2,2,2-trifluoroethyl)-1H-indol-2-yl}prop-2-yn-1-yl)amino]pyridin-2-yl}propanenitrile, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Abendroth, J, Lorimer, D.D, Vu, B, Takana, N. | | Deposit date: | 2024-05-10 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Rezatapopt (PC14586), a First-in-Class, Small-Molecule Reactivator of p53 Y220C Mutant in Development.
Acs Med.Chem.Lett., 16, 2025
|
|
8OXB
 
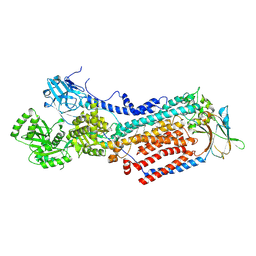 | | Cryo-EM structure of ATP8B1-CDC50A in E2-Pi conformation with occluded PC | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
1MCP
 
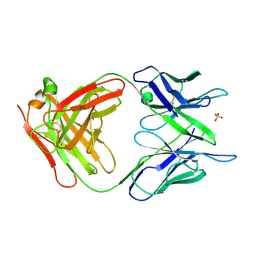 | | PHOSPHOCHOLINE BINDING IMMUNOGLOBULIN FAB MC/PC603. AN X-RAY DIFFRACTION STUDY AT 2.7 ANGSTROMS | | Descriptor: | IGA-KAPPA MCPC603 FAB (HEAVY CHAIN), IGA-KAPPA MCPC603 FAB (LIGHT CHAIN), SULFATE ION | | Authors: | Satow, Y, Cohen, G.H, Padlan, E.A, Davies, D.R. | | Deposit date: | 1984-07-09 | | Release date: | 1985-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A.
J.Mol.Biol., 190, 1986
|
|
8I42
 
 | |
8I47
 
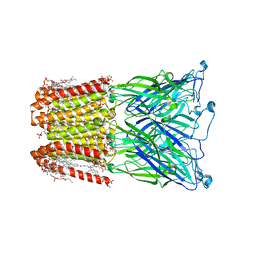 | |
2X6C
 
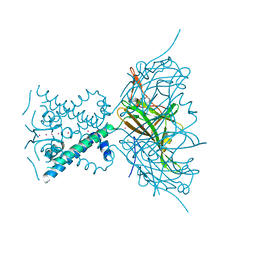 | | Potassium Channel from Magnetospirillum Magnetotacticum | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, CHLORIDE ION, PHOSPHOCHOLINE, ... | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, Vandenberg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
2X6A
 
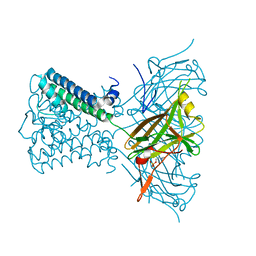 | | Potassium Channel from Magnetospirillum Magnetotacticum | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, PHOSPHOCHOLINE, POTASSIUM ION | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, Vandenberg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
2X6B
 
 | | Potassium Channel from Magnetospirillum Magnetotacticum | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, BARIUM ION, PHOSPHOCHOLINE, ... | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, Vandenberg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
8BEL
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII membrane domain) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEH
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane tip) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEF
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
3LKF
 
 | | LEUKOCIDIN F (HLGB) FROM STAPHYLOCOCCUS AUREUS WITH PHOSPHOCHOLINE BOUND | | Descriptor: | LEUKOCIDIN F SUBUNIT, PHOSPHOCHOLINE | | Authors: | Olson, R, Nariya, H, Yokota, K, Kamio, Y, Gouaux, J.E. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel.
Nat.Struct.Biol., 6, 1999
|
|
6AKP
 
 | | Crystal Structural of FOXC2 DNA binding domain bound to PC promoter | | Descriptor: | DNA (5'-D(AP*CP*AP*CP*AP*AP*AP*TP*AP*TP*TP*TP*GP*TP*GP*T)-3'), Forkhead box protein C2, MAGNESIUM ION | | Authors: | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
2BIB
 
 | | Crystal structure of the complete modular teichioic acid phosphorylcholine esterase Pce (CbpE) from Streptococcus pneumoniae | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, PHOSPHOCHOLINE, ... | | Authors: | Hermoso, J.A, Lagartera, L, Gonzalez, A, Garcia, P, Martinez-Ripoll, M, Garcia, J.L, Menendez, M. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights Into Pneumococcal Pathogenesis from Crystal Structure of the Modular Teichoic Acid Phosphorylcholine Esterase Pce
Nat.Struct.Mol.Biol., 12, 2005
|
|
