8Y6O
 
 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U11 and tri-snRNP part | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Centrosomal AT-AC splicing factor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
8SCZ
 
 | |
8SD0
 
 | |
8VXY
 
 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | Authors: | Tuck, O.T, Hu, J.J, Doudna, J.A. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
8VXA
 
 | |
8VX9
 
 | |
8VXC
 
 | |
8G9U
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | CRISPR-associated protein, Csd1 family, Csd2 family, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8OZ0
 
 | | Structure of a human 48S translation initiation complex with eIF4F and eIF4A | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Fraser, C.S, Ramakrishnan, V. | | Deposit date: | 2023-05-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of a human translation initiation complex reveals two independent roles for the helicase eIF4A.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IJU
 
 | | ATP-dependent RNA helicase DDX39A (URH49delta41) | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DDX39A, PHOSPHATE ION, ... | | Authors: | Mikami, B, Fujita, K, Masuda, S, Kojima, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural differences between the closely related RNA helicases, UAP56 and URH49, fashion distinct functional apo-complexes.
Nat Commun, 15, 2024
|
|
8QO9
 
 | | Cryo-EM structure of a human spliceosomal B complex protomer | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, MINX pre-mRNA, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (5.29 Å) | | Cite: | Cryo-EM analyses of dimerized spliceosomes provide new insights into the functions of B complex proteins.
Embo J., 43, 2024
|
|
8QCF
 
 | | yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Antiviral helicase SKI2, Exosome complex component CSL4, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
7YMF
 
 | | Crystal Structure of DDX3X449_450ET>DP | | Descriptor: | ATP-dependent RNA helicase DDX3X | | Authors: | Xiong, J. | | Deposit date: | 2022-07-28 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of DDX3X449_450ET>DP
To Be Published
|
|
8Q9T
 
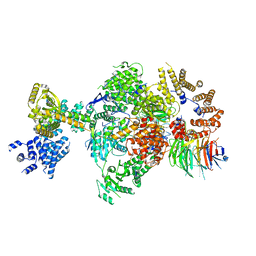 | | CryoEM structure of a S. Cerevisiae Ski238 complex bound to RNA | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, RNA (5'-R(P*UP*UP*UP*U)-3'), ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8PJJ
 
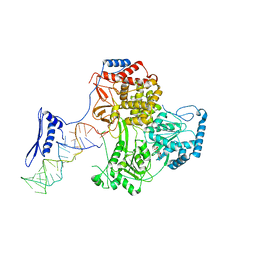 | |
8QCA
 
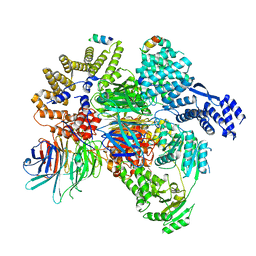 | | CryoEM structure of a S. Cerevisiae Ski2387 complex in the closed state bound to RNA | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, RNA (5'-R(P*UP*UP*UP*U)-3'), ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8G7U
 
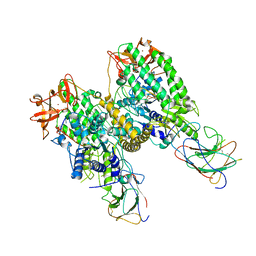 | |
8G7T
 
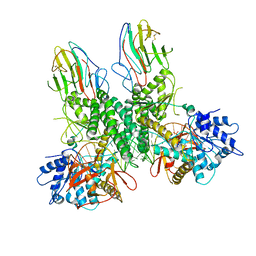 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-end) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
8G7V
 
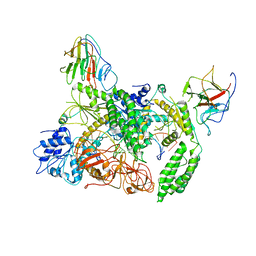 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-inter) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
8QCB
 
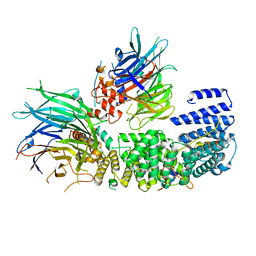 | | CryoEM structure of a S. Cerevisiae Ski2387 complex in the open state | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, Superkiller protein 3, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8PJB
 
 | |
8B9L
 
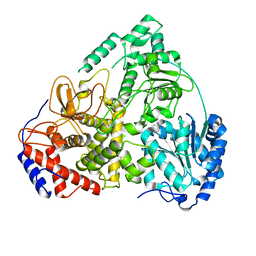 | | Cryo-EM structure of MLE | | Descriptor: | Dosage compensation regulator | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
8B9K
 
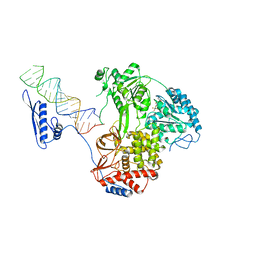 | |
8B9J
 
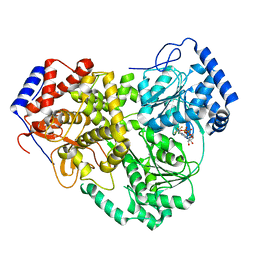 | | Cryo-EM structure of MLE in complex with ADP:AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dosage compensation regulator, MAGNESIUM ION, ... | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
8B9I
 
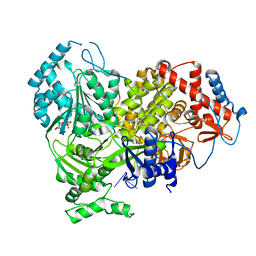 | |
