5Q16
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-{2-[(4-chloro-2-methylphenoxy)methyl]-6-fluoro-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q19
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-[2-(2,4-dimethoxyphenyl)-1H-benzimidazol-1-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0R
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,1-dibenzyl-6-[(2-fluorophenyl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0Z
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, ethyl (5S)-3-(3,4-difluorobenzene-1-carbonyl)-1,1-dimethyl-1,2,3,4,5,6-hexahydroazepino[4,5-b]indole-5-carboxylate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 3-(2-chlorophenyl)-N-[(1R)-1-(naphthalen-2-yl)ethyl]-5-(propan-2-yl)-1,2-oxazole-4-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
6LQR
 
 | |
6LQV
 
 | |
6LQT
 
 | |
6LQQ
 
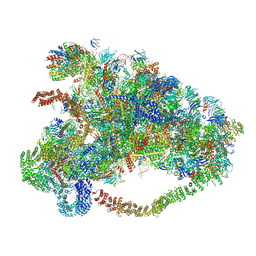 | |
6LQP
 
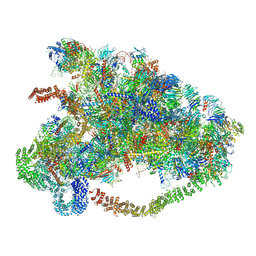 | |
8T1L
 
 | | Atomic model of the mammalian mouse Mediator complex with CKM module | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.83 Å) | | Cite: | A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers.
To Be Published
|
|
5T0I
 
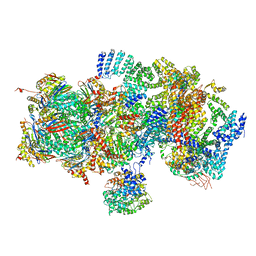 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6LUA
 
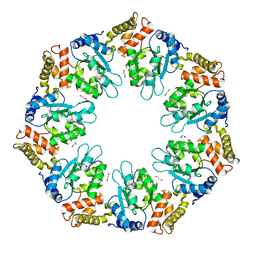 | |
5T0C
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-15 | | Release date: | 2016-10-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0J
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5Q0O
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-{2-[4-(benzenecarbonyl)phenyl]-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, CHLORIDE ION, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q14
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}-3-fluorobenzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1E
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-bromo-1-{[4-(1H-tetrazol-5-yl)phenyl]methyl}-1'-(thiophene-2-sulfonyl)spiro[indole-3,4'-piperidin]-2(1H)-one, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5SXP
 
 | | STRUCTURAL BASIS FOR THE INTERACTION BETWEEN ITCH PRR AND BETA-PIX | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Rho guanine nucleotide exchange factor 7 | | Authors: | Cappadocia, L, Desrochers, G, Lussier-Price, M, Angers, A, Omichinski, J.G. | | Deposit date: | 2016-08-09 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of interactions between SH3 domain-containing proteins and the proline-rich region of the ubiquitin ligase Itch.
J. Biol. Chem., 292, 2017
|
|
6LQU
 
 | |
5T0G
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0H
 
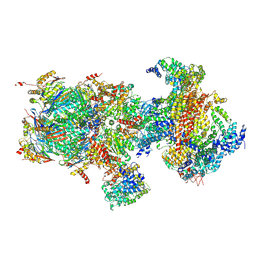 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
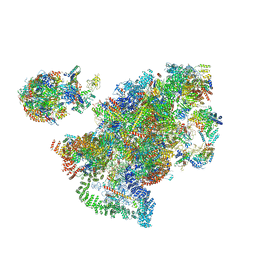
6LQS
 
 | |
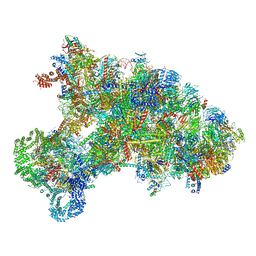
6KE6
 
 | |
5TJ8
 
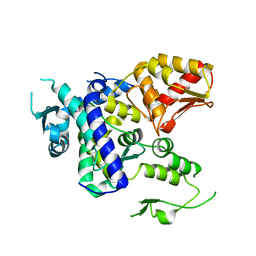 | |
