1QIA
 
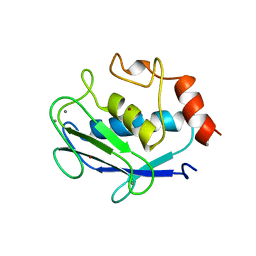 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, STROMELYSIN-1, ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
5X1X
 
 | |
5XIN
 
 | |
3KB8
 
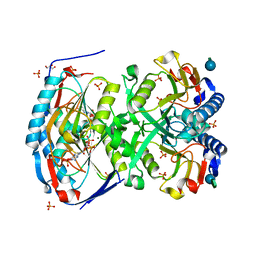 | | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP
To be Published
|
|
5X2U
 
 | |
1QPU
 
 | | SOLUTION STRUCTURE OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Faraone-Mennella, J, Rosato, A, Barker, P.D, Fersht, A.R. | | Deposit date: | 1999-05-30 | | Release date: | 1999-06-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized Escherichia coli cytochrome b562.
Biochemistry, 38, 1999
|
|
3KCG
 
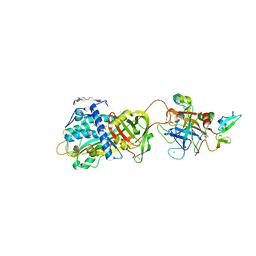 | | Crystal structure of the antithrombin-factor IXa-pentasaccharide complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, Antithrombin-III, ... | | Authors: | Huntington, J.A, Johnson, D.J.D. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of factor IXa recognition by heparin-activated antithrombin revealed by a 1.7-A structure of the ternary complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6I6B
 
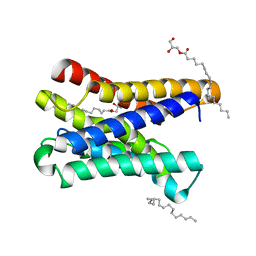 | |
5X3I
 
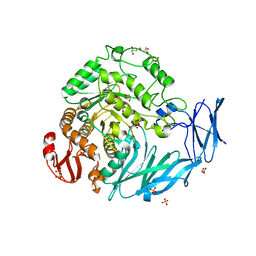 | | Kfla1895 D451A mutant | | Descriptor: | GLYCEROL, Glycoside hydrolase family 31, SULFATE ION | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycoside hydrolase mutant
To Be Published
|
|
1QQL
 
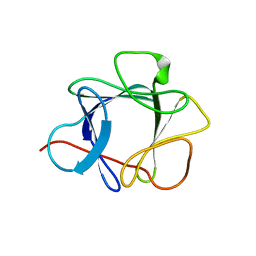 | | THE CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 7/1 CHIMERA | | Descriptor: | FIBROBLAST GROWTH FACTOR 7/1 CHIMERA | | Authors: | Ye, S, Luo, Y, Pelletier, H, McKeehan, W.L. | | Deposit date: | 1999-06-07 | | Release date: | 2000-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs.
Biochemistry, 40, 2001
|
|
1QPV
 
 | | YEAST COFILIN | | Descriptor: | YEAST COFILIN | | Authors: | Fedorov, A.A, Lappalainen, P, Fedorov, E.V, Drubin, D.G, Almo, S.C. | | Deposit date: | 1999-05-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of yeast cofilin.
Nat.Struct.Biol., 4, 1997
|
|
6IR4
 
 | | Crystal structure of BioU from Synechocystis sp.PCC6803 (apo form) | | Descriptor: | Slr0355 protein | | Authors: | Sakaki, K, Oishi, K, Shimizu, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2018-11-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
1QQ2
 
 | | CRYSTAL STRUCTURE OF A MAMMALIAN 2-CYS PEROXIREDOXIN, HBP23. | | Descriptor: | CHLORIDE ION, THIOREDOXIN PEROXIDASE 2 | | Authors: | Hirotsu, S, Abe, Y, Okada, K, Nagahara, N, Hori, H, Nishino, T, Hakoshima, T. | | Deposit date: | 1999-06-10 | | Release date: | 1999-10-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6I73
 
 | |
1QQV
 
 | |
5X4L
 
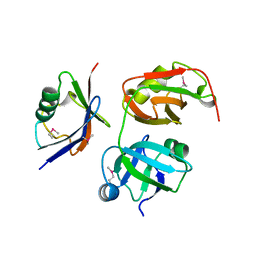 | | Crystal structure of the UBX domain of human UBXD7 in complex with p97 N domain | | Descriptor: | Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 7 | | Authors: | Jiang, T, Li, Z, Wang, Y, Xu, M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structures of the UBX domain of human UBXD7 and its complex with p97 ATPase
Biochem. Biophys. Res. Commun., 486, 2017
|
|
1QR1
 
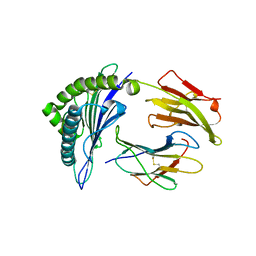 | | POOR BINDING OF A HER-2/NEU EPITOPE (GP2) TO HLA-A2.1 IS DUE TO A LACK OF INTERACTIONS IN THE CENTER OF THE PEPTIDE | | Descriptor: | BETA-2 MICROGLOBULIN, GP2 PEPTIDE, HLA-A2.1 HEAVY CHAIN | | Authors: | Kuhns, J.J, Batalia, M.A, Yan, S, Collins, E.J. | | Deposit date: | 1999-06-17 | | Release date: | 2000-01-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Poor binding of a HER-2/neu epitope (GP2) to HLA-A2.1 is due to a lack of interactions with the center of the peptide.
J.Biol.Chem., 274, 1999
|
|
6I7U
 
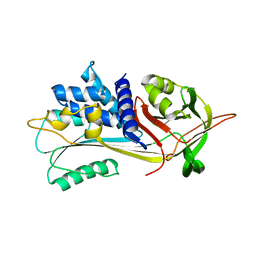 | |
5X3D
 
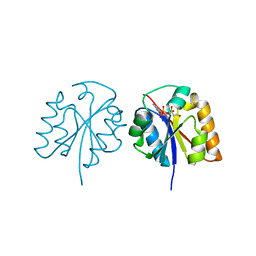 | | Crystal structure of HEP-CMP-bound form of cytidylyltransferase (CyTase) domain of Fom1 from Streptomyces wedmorensis | | Descriptor: | Phosphoenolpyruvate phosphomutase, [[(2R,3S,4R,5R)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-(2-hydroxyethyl)phosphinic acid | | Authors: | Tomita, T, Cho, S.H, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2017-02-04 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fosfomycin Biosynthesis via Transient Cytidylylation of 2-Hydroxyethylphosphonate by the Bifunctional Fom1 Enzyme
ACS Chem. Biol., 12, 2017
|
|
1QQI
 
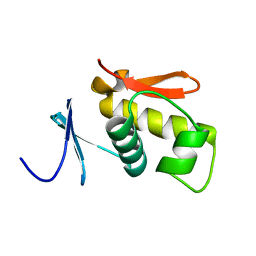 | | SOLUTION STRUCTURE OF THE DNA-BINDING AND TRANSACTIVATION DOMAIN OF PHOB FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Okamura, H, Hanaoka, S, Nagadoi, A, Makino, K, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.
J.Mol.Biol., 295, 2000
|
|
1QQR
 
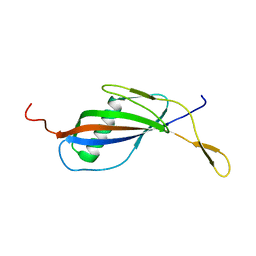 | | CRYSTAL STRUCTURE OF STREPTOKINASE DOMAIN B | | Descriptor: | STREPTOKINASE DOMAIN B | | Authors: | Spraggon, G, Zhang, X.X, Ponting, C.P, Fox, V.F, Phillips, C, Smith, R.A.G, Jones, E.Y, Dobson, C, Stuart, D.I. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Streptokinse Domain B
To be Published
|
|
5X4K
 
 | | The complex crystal structure of Pyrococcus furiosus RecJ and CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Uncharacterized protein, ZINC ION | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
1QRA
 
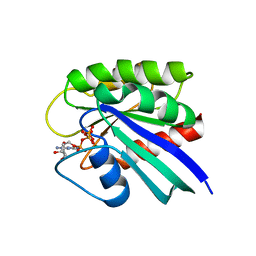 | | STRUCTURE OF P21RAS IN COMPLEX WITH GTP AT 100 K | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Scheidig, A, Burmester, C, Goody, R.S. | | Deposit date: | 1999-06-12 | | Release date: | 1999-11-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins.
Structure Fold.Des., 7, 1999
|
|
6I8J
 
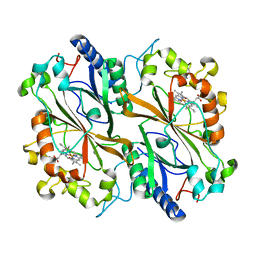 | | Dye type peroxidase Aa from Streptomyces lividans: 131.2 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-11-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
1QOB
 
 | | FERREDOXIN MUTATION D62K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
