8JFS
 
 | | Phosphate bound acylphosphatase from Deinococcus radiodurans at 1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Acylphosphatase, CITRIC ACID, ... | | Authors: | Khakerwala, Z, Kumar, A, Makde, R.D. | | Deposit date: | 2023-05-18 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of phosphate bound Acyl phosphatase mini-enzyme from Deinococcus radiodurans at 1 angstrom resolution.
Biochem.Biophys.Res.Commun., 671, 2023
|
|
8IU9
 
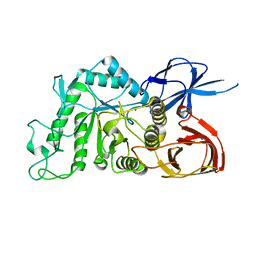 | |
8IUB
 
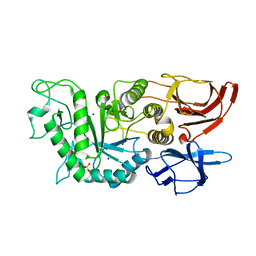 | |
7N16
 
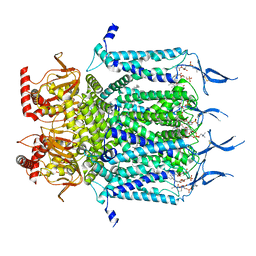 | | Structure of TAX-4_R421W apo closed state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
8FHA
 
 | |
8J5A
 
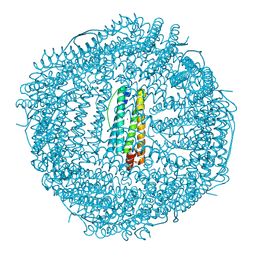 | | Single-particle cryo-EM structure of mouse apoferritin at 1.19 Angstrom resolution (Dataset A) | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Kawakami, K, Maki-Yonekura, S, Hamaguchi, T, Takaba, K, Yonekura, K. | | Deposit date: | 2023-04-21 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (1.19 Å) | | Cite: | Measurement of charges and chemical bonding in a cryo-EM structure.
Commun Chem, 6, 2023
|
|
4HZY
 
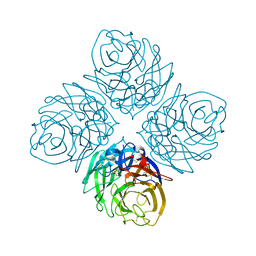 | | Crystal structure of influenza A neuraminidase N3-H274Y | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
8GYR
 
 | | Crystal structure of a variable region segment of Leptospira host-interacting outer surface protein, LigA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kumar, P, Akif, M. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of a variable region segment of Leptospira host-interacting outer surface protein, LigA, reveals the orientation of Ig-like domains.
Int.J.Biol.Macromol., 244, 2023
|
|
7MZ2
 
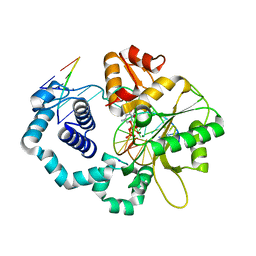 | |
7MZ1
 
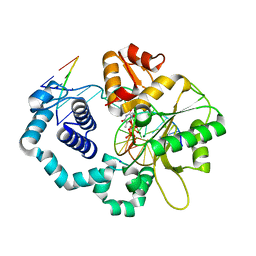 | |
7MZ0
 
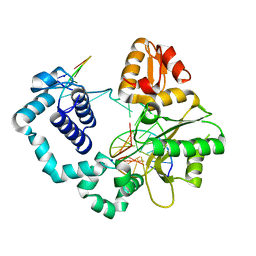 | |
7MZ4
 
 | |
7MZ8
 
 | |
7MZ3
 
 | |
7N4P
 
 | | Crystal Structure of Lizard Cadherin-23 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin 23, SODIUM ION | | Authors: | Nisler, C.R, Sotomayor, M. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Interpreting the Evolutionary Echoes of a Protein Complex Essential for Inner-Ear Mechanosensation.
Mol.Biol.Evol., 40, 2023
|
|
8GN9
 
 | | SPFH domain of Pyrococcus horikoshii stomatin | | Descriptor: | SODIUM ION, Stomatin homolog PH1511 | | Authors: | Komatsu, T, Matsui, I, Yokoyama, H. | | Deposit date: | 2022-08-23 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mutational studies suggest key residues to determine whether stomatin SPFH domains form dimers or trimers.
Biochem Biophys Rep, 32, 2022
|
|
8J6G
 
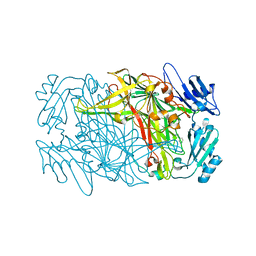 | |
8GPZ
 
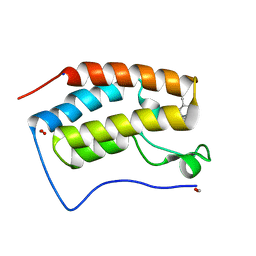 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with C239-0012 | | Descriptor: | 3-methyl-6-(4-methylpiperidin-1-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4, FORMIC ACID, ... | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2022-08-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
8GQ0
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with STL233497 | | Descriptor: | Bromodomain-containing protein 4, FORMIC ACID, GLYCEROL, ... | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2022-08-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
7N4K
 
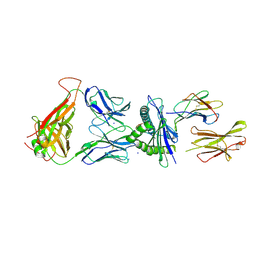 | | 6218 TCR in complex with H2-Db PA 224 | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 and T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5C
 
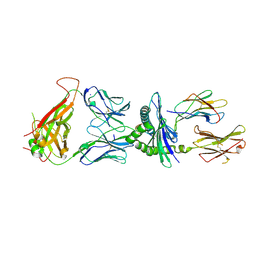 | | 6218 TCR in complex with H2Db PA with an engineered TCR-pMHC disulfide bond | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 with T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-05 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5P
 
 | | 6218 TCR in complex with H2-Db PA224-233 with a cysteine mutant | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 and T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5Q
 
 | |
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
7NBB
 
 | |
