3LC9
 
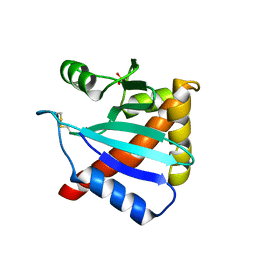 | |
4GN3
 
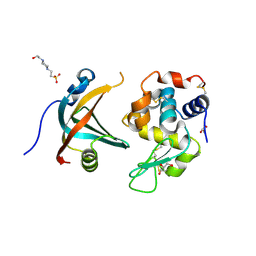 | | OBody AM1L10 bound to hen egg-white lysozyme | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Lysozyme C, ... | | Authors: | Steemson, J.D, Liddament, M.T. | | Deposit date: | 2012-08-16 | | Release date: | 2013-08-21 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tracking Molecular Recognition at the Atomic Level with a New Protein Scaffold Based on the OB-Fold.
Plos One, 9, 2014
|
|
3UV9
 
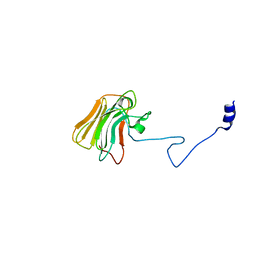 | | Structure of the rhesus monkey TRIM5alpha deltav1 PRYSPRY domain | | Descriptor: | Tripartite motif-containing protein 5 | | Authors: | Biris, N, Yang, Y, Taylor, A.B, Tomashevskii, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D. | | Deposit date: | 2011-11-29 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5EWN
 
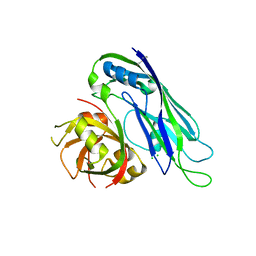 | | Crystal structure of the human astrovirus 1 capsid protein core domain at 2.6 A resolution | | Descriptor: | CHLORIDE ION, Structural protein | | Authors: | York, R.L, Yousefi, P.A, Bogdanoff, W, Haile, S, Tripathi, S, DuBois, R.M. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-23 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structural, Mechanistic, and Antigenic Characterization of the Human Astrovirus Capsid.
J.Virol., 90, 2015
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5J1A
 
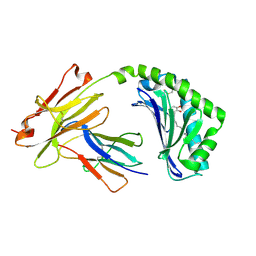 | | Antigen presenting molecule | | Descriptor: | 3-[(8Z,11Z)-pentadeca-8,11-dien-1-yl]benzene-1,2-diol, Beta-2-microglobulin, T-cell surface glycoprotein CD1a | | Authors: | Yongqing, T, Rossjohn, J, Le Nours, J, Marquez, E, Winau, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | CD1a on Langerhans cells controls inflammatory skin disease.
Nat. Immunol., 17, 2016
|
|
6IWY
 
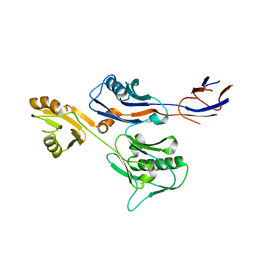 | |
5XIX
 
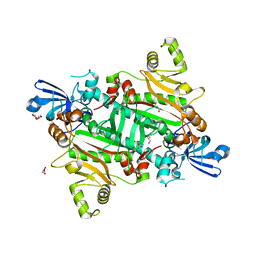 | | The canonical domain of human asparaginyl-tRNA synthetase | | Descriptor: | Asparagine--tRNA ligase, cytoplasmic, GLYCEROL | | Authors: | Park, J.S, Han, B.W. | | Deposit date: | 2017-04-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Unique N-terminal extension domain of human asparaginyl-tRNA synthetase elicits CCR3-mediated chemokine activity.
Int. J. Biol. Macromol., 120, 2018
|
|
4GJX
 
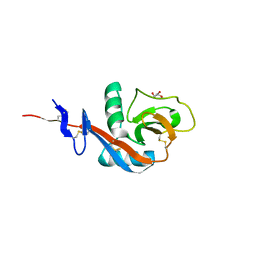 | |
5IZ7
 
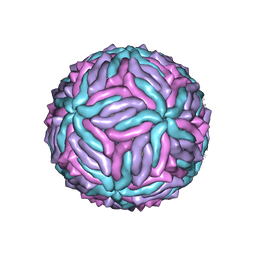 | | Cryo-EM structure of thermally stable Zika virus strain H/PF/2013 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, structural protein E, structural protein M | | Authors: | Kostyuchenko, V.A, Zhang, S, Fibriansah, G, Lok, S.M. | | Deposit date: | 2016-03-25 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the thermally stable Zika virus
Nature, 533, 2016
|
|
4GLV
 
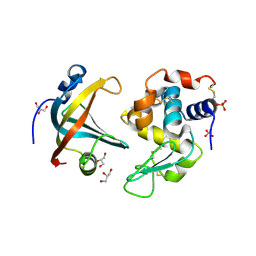 | | OBody AM3L09 bound to hen egg-white lysozyme | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Lysozyme C, ... | | Authors: | Steemson, J.D. | | Deposit date: | 2012-08-15 | | Release date: | 2013-08-21 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Tracking Molecular Recognition at the Atomic Level with a New Protein Scaffold Based on the OB-Fold.
Plos One, 9, 2014
|
|
3PHS
 
 | |
8K3K
 
 | |
5XXZ
 
 | |
5IUS
 
 | | Crystal structure of human PD-L1 in complex with high affinity PD-1 mutant | | Descriptor: | CHLORIDE ION, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Pascolutti, R, Sun, X, Kao, J, Maute, R, Ring, A.M, Bowman, G.R, Kruse, A.C. | | Deposit date: | 2016-03-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Structure and Dynamics of PD-L1 and an Ultra-High-Affinity PD-1 Receptor Mutant.
Structure, 24, 2016
|
|
1C78
 
 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
5JJ4
 
 | | Crystal Structure of a Variant Human Activation-induced Deoxycytidine Deaminase as an MBP fusion protein | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Single-stranded DNA cytosine deaminase, ZINC ION, ... | | Authors: | Pedersen, L.C, Goodman, M.F, Pham, P, Afif, S.A. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Structural analysis of the activation-induced deoxycytidine deaminase required in immunoglobulin diversification.
DNA Repair (Amst.), 43, 2016
|
|
1NAK
 
 | | IGG1 FAB FRAGMENT (83.1) COMPLEX WITH 16-RESIDUE PEPTIDE (RESIDUES 304-321 OF HIV-1 GP120 (MN ISOLATE)) | | Descriptor: | Fab 83.1 - heavy chain, Fab 83.1 - light chain, Peptide MP1 | | Authors: | Stanfield, R.L, Ghiara, J.B, Saphire, E.O, Profy, A.T, Wilson, I.A. | | Deposit date: | 2002-11-27 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Recurring conformation of the human immunodeficiency virus type 1 gp120 V3 loop.
Virology, 315, 2003
|
|
1C77
 
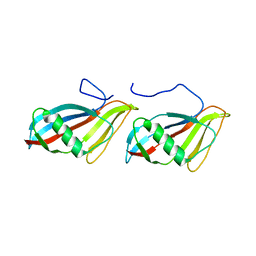 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1CCZ
 
 | | CRYSTAL STRUCTURE OF THE CD2-BINDING DOMAIN OF CD58 (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3) AT 1.8-A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CD58) | | Authors: | Ikemizu, S, Sparks, L.M, Van Der Merwe, P.A, Harlos, K, Stuart, D.I, Jones, E.Y, Davis, S.J. | | Deposit date: | 1999-03-02 | | Release date: | 1999-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the CD2-binding domain of CD58 (lymphocyte function-associated antigen 3) at 1.8-A resolution.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1GX0
 
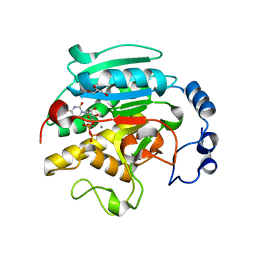 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - BETA-D-GALACTOSE COMPLEX | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
5FEG
 
 | |
1C79
 
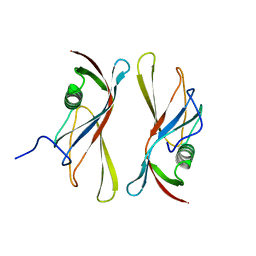 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
6HBU
 
 | | Cryo-EM structure of the ABCG2 E211Q mutant bound to ATP and Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 2, MAGNESIUM ION | | Authors: | Manolaridis, I, Jackson, S.M, Taylor, N.M.I, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-07 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states.
Nature, 563, 2018
|
|
6HCO
 
 | | Cryo-EM structure of the ABCG2 E211Q mutant bound to estrone 3-sulfate and 5D3-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Manolaridis, I, Jackson, S.M, Taylor, N.M.I, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states.
Nature, 563, 2018
|
|
