6Y2S
 
 | |
4ILU
 
 | | Crystal structure of Mycobacterium tuberculosis CarD | | Descriptor: | RNA polymerase-binding transcription factor CarD, SULFATE ION | | Authors: | Thakur, K.G, Kaur, G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis CarD, an essential RNA polymerase binding protein, reveals a quasidomain-swapped dimeric structural architecture.
Proteins, 82, 2014
|
|
6Y2R
 
 | |
2N4L
 
 | | Solution Structure of the HIV-1 Intron Splicing Silencer and its Interactions with the UP1 Domain of hnRNP A1 | | Descriptor: | RNA (53-MER) | | Authors: | Tolbert, B.S, Jain, N, Morgan, C.E, Rife, B.D, Salemi, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the HIV-1 Intron Splicing Silencer and Its Interactions with the UP1 Domain of Heterogeneous Nuclear Ribonucleoprotein (hnRNP) A1.
J.Biol.Chem., 291, 2016
|
|
3WHO
 
 | | X-ray-Crystallographic Structure of an RNase Po1 Exhibiting Anti-tumor Activity | | Descriptor: | Guanyl-specific ribonuclease Po1 | | Authors: | Kobayashi, H, Katsurtani, T, Hara, Y, Motoyoshi, N, Itagaki, T, Akita, F, Higashiura, A, Yamada, Y, Suzuki, M, Inokuchi, N. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystallographic structure of RNase Po1 that exhibits anti-tumor activity.
Biol.Pharm.Bull., 37, 2014
|
|
3CLJ
 
 | | Structure of the RNA polymerase II CTD-interacting domain of Nrd1 | | Descriptor: | GLYCEROL, Protein NRD1, SULFATE ION | | Authors: | Vasiljeva, L, Kim, M, Mutschler, H, Buratowski, S, Meinhart, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2CPE
 
 | | Solution structure of the RNA recognition motif of Ewing Sarcoma(EWS) protein | | Descriptor: | RNA-binding protein EWS | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA recognition motif of Ewing Sarcoma(EWS) protein
To be Published
|
|
4DK3
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4DKA
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, SODIUM ION, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
6EZ4
 
 | | NMR structure of the C-terminal domain of the human RPAP3 protein | | Descriptor: | RNA polymerase II-associated protein 3 | | Authors: | Fabre, P, Chagot, M.E, Bragantini, B, Manival, X, Quinternet, M. | | Deposit date: | 2017-11-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones.
Nat Commun, 9, 2018
|
|
1FHT
 
 | | RNA-BINDING DOMAIN OF THE U1A SPLICEOSOMAL PROTEIN U1A117, NMR, 43 STRUCTURES | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1996-02-21 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding.
J.Mol.Biol., 257, 1996
|
|
4DK6
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
8K8B
 
 | |
5MRX
 
 | |
418D
 
 | |
2DHG
 
 | | Solution structure of the C-terminal RNA recognition motif in tRNA selenocysteine associated protein | | Descriptor: | tRNA selenocysteine associated protein (SECP43) | | Authors: | Imai, T, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-23 | | Release date: | 2006-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal RNA recognition motif in tRNA selenocysteine associated protein
To be Published
|
|
2KDV
 
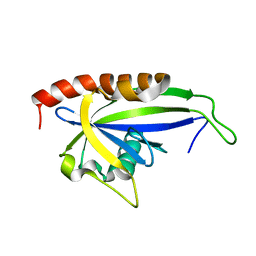 | |
2R7O
 
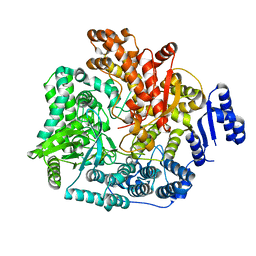 | | Crystal Structure of VP1 apoenzyme of Rotavirus SA11 (N-terminal hexahistidine-tagged) | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-09 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
3N4K
 
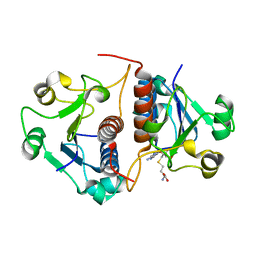 | | Putative RNA methyltransferase from Yersinia pestis in complex with S-ADENOSYL-L-HOMOCYSTEINE. | | Descriptor: | RNA methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Osipiuk, J, Maltseva, N, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray crystal structure of putative RNA methyltransferase from Yersinia pestis.
To be Published
|
|
2R7Q
 
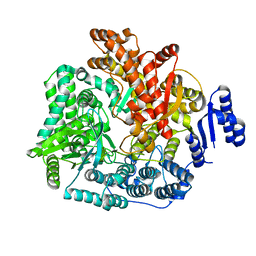 | | Crystal Structure of VP1 apoenzyme of Rotavirus SA11 (C-terminal hexahistidine-tagged) | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-09 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2K9L
 
 | | Structure of the Core Binding Domain of sigma54 | | Descriptor: | RNA polymerase sigma factor RpoN | | Authors: | Hong, E, Wemmer, D. | | Deposit date: | 2008-10-19 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the RNA polymerase core-binding domain of sigma(54) reveals a likely conformational fracture point
J.Mol.Biol., 390, 2009
|
|
3N4J
 
 | | Putative RNA methyltransferase from Yersinia pestis | | Descriptor: | RNA methyltransferase, SULFATE ION | | Authors: | Osipiuk, J, Maltseva, N, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystal structure of putative RNA methyltransferase from Yersinia pestis.
To be Published
|
|
4NJ5
 
 | | Crystal structure of SUVH9 | | Descriptor: | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH9, ZINC ION | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.
Nature, 507, 2014
|
|
2XHU
 
 | | HCV-J4 NS5B Polymerase Orthorhombic Crystal Form | | Descriptor: | RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
1WF1
 
 | | Solution structure of RRM domain in RNA-binding protein NP_057951 | | Descriptor: | RNA-binding protein Raly | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in RNA-binding protein NP_057951
To be Published
|
|
