6PV5
 
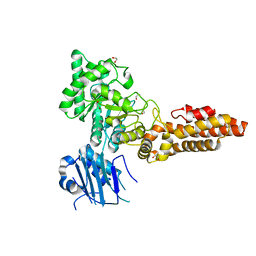 | | Structure of CpGH84B | | Descriptor: | 1,2-ETHANEDIOL, Putative O-GlcNAcase nagJ | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens.
Glycobiology, 30, 2019
|
|
3WYX
 
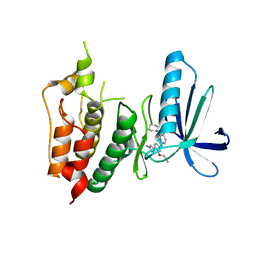 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 6-((3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-2-(cyclohexylamino)nicotinonitrile | | Descriptor: | 6-{[3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl]amino}-2-(cyclohexylamino)pyridine-3-carbonitrile, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A unique hinge binder of extremely selective aminopyridine-based Mps1 (TTK) kinase inhibitors with cellular activity.
Bioorg.Med.Chem., 23, 2015
|
|
3WZK
 
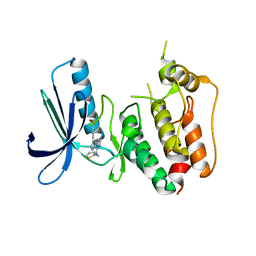 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH N-cyclopropyl-4-(8-((thiophen-2-ylmethyl)amino)imidazo[1,2-a]pyrazin-3-yl)benzamide | | Descriptor: | CHLORIDE ION, Dual specificity protein kinase TTK, N-cyclopropyl-4-{8-[(thiophen-2-ylmethyl)amino]imidazo[1,2-a]pyrazin-3-yl}benzamide | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of imidazo[1,2-b]pyridazine derivatives: selective and orally available Mps1 (TTK) kinase inhibitors exhibiting remarkable antiproliferative activity.
J.Med.Chem., 58, 2015
|
|
6PU3
 
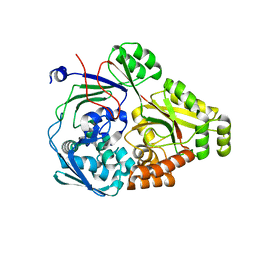 | | ABC transporter-associated periplasmic binding protein DppA from Helicobacter pylori | | Descriptor: | Heme-binding protein A, SER-THR-SER-ALA | | Authors: | Rahman, M.M, Machuca, M.A, Khan, M.F, Barlow, C.K, Schittenhelm, R.B, Roujeinikova, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori.
J.Bacteriol., 201, 2019
|
|
5MGF
 
 | |
7QOF
 
 | | Icosahedral capsid of the phicrAss001 virion | | Descriptor: | Auxiliary capsid protein gp36, Head fiber dimer protein gp29, Head fiber trimer protein gp21, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOG
 
 | | Portal protein assembly of the phicrAss001 virion with C12 symmetry imposed | | Descriptor: | Cargo protein 1 gp45, Portal protein gp20, Ring protein 1 gp43, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOH
 
 | | Unique vertex of the phicrAss001 virion with C5 symmetry imposed | | Descriptor: | Auxiliary capsid protein gp36, Head fiber trimer protein gp21, MAGNESIUM ION, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOK
 
 | | Tail muzzle assembly of the phicrAss001 virion with C6 symmetry imposed | | Descriptor: | MAGNESIUM ION, Muzzle bound helix, Muzzle protein gp44, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOJ
 
 | | Tail barrel assembly of the phicrAss001 virion with C12 symmetry imposed | | Descriptor: | Cargo protein 1 gp45, MAGNESIUM ION, Portal protein gp20, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOL
 
 | | Tail assembly of the phicrAss001 virion with C6 symmetry imposed | | Descriptor: | Cargo protein 1 gp45, MAGNESIUM ION, Muzzle bound helix, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7UOA
 
 | | MAGEA4-MTP1 linear peptide complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MTP-1, ... | | Authors: | Williams, R.S, Tumbale, P.S. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery and Structural Basis of the Selectivity of Potent Cyclic Peptide Inhibitors of MAGE-A4.
J.Med.Chem., 65, 2022
|
|
7QOI
 
 | | Unique vertex of the phicrAss001 virion | | Descriptor: | Auxiliary capsid protein gp36, Cargo protein 1 gp45, Head fiber trimer protein gp21, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
6PWI
 
 | | Structure of CpGH84D | | Descriptor: | 1,2-ETHANEDIOL, Putative hyaluronoglucosaminidase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens.
Glycobiology, 30, 2019
|
|
7UAY
 
 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-{[(pyridin-3-yl)methyl]sulfanyl}propanamide)bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Ir(III)-Based Agents for Monitoring the Cytochrome P450 3A4 Active Site Occupancy.
Inorg.Chem., 61, 2022
|
|
7UAZ
 
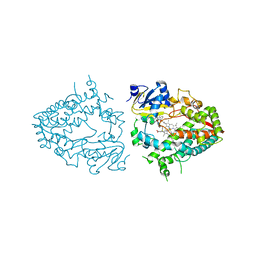 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-{[(pyridin-3-yl)methyl]sulfanyl}propanamide)bis[2-(pyridin-2-yl-kappaN)phenyl-kappaC~1~]iridium, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Ir(III)-Based Agents for Monitoring the Cytochrome P450 3A4 Active Site Occupancy.
Inorg.Chem., 61, 2022
|
|
6PXZ
 
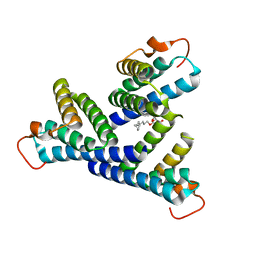 | |
7UQR
 
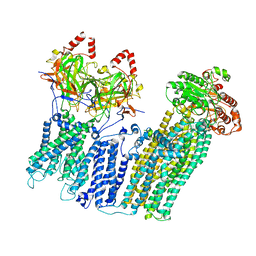 | |
6W13
 
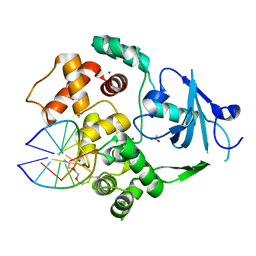 | |
3ZL1
 
 | | A thiazolyl-mannoside bound to FimH, monoclinic space group | | Descriptor: | CHLORIDE ION, N-{5-[(1R)-1-hydroxyethyl]-1,3-thiazol-2-yl}-alpha-D-mannopyranosylamine, PROTEIN FIMH | | Authors: | Brument, S, Sivignon, A, Dumych, T.I, Moreau, N, Roos, G, Guerardel, Y, Chalopin, T, Deniaud, D, Bilyy, R.O, Darfeuille-Michaud, A, Bouckaert, J, Gouin, S.G. | | Deposit date: | 2013-01-27 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Thiazolylaminomannosides as Potent Antiadhesives of Type 1 Piliated Escherichia Coli Isolated from Crohn'S Disease Patients.
J.Med.Chem., 56, 2013
|
|
6W3R
 
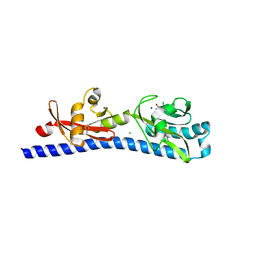 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with 3-methylisoleucine | | Descriptor: | 3-methyl-L-alloisoleucine, CHLORIDE ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6PZA
 
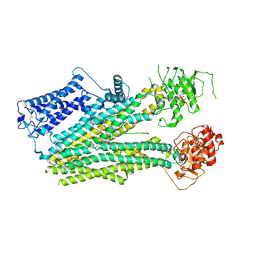 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
6Q6L
 
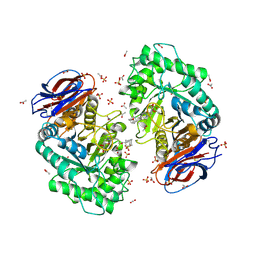 | | Crystal structure of recombinant human beta-glucocerebrosidase in complex with adamantyl-cyclophellitol inhibitor (ME656) | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},5~{S})-4-[[4-[3-(1-adamantylmethoxy)propyl]-1,2,3-triazol-1-yl]methyl]cyclohexane-1,2,3,5-tetrol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Functionalized Cyclophellitols Are Selective Glucocerebrosidase Inhibitors and Induce a Bona Fide Neuropathic Gaucher Model in Zebrafish.
J.Am.Chem.Soc., 141, 2019
|
|
6W3X
 
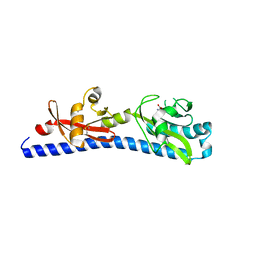 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-valine | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, SULFATE ION, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
7RNJ
 
 | | S2P6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | Monoclonal antibody S2P6 Fab heavy chain, Monoclonal antibody S2P6 Fab light chain, SULFATE ION, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M, Sauer, M.M, Veesler, D. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Broad betacoronavirus neutralization by a stem helix-specific human antibody.
Science, 373, 2021
|
|
