1HEG
 
 | | The crystal structures at 2.2 angstroms resolution of hydroxyethylene-based inhibitors bound to human immunodeficiency virus type 1 protease show that the inhibitors are present in two distinct orientations | | Descriptor: | HIV-1 PROTEASE, methyl N-{(4S,5S)-5-[(L-alanyl-L-alanyl)amino]-4-hydroxy-6-phenylhexanoyl}-L-valyl-L-valinate | | Authors: | Murthy, K, Winborne, E.L, Minnich, M.D, Culp, J.S, Debouck, C. | | Deposit date: | 1992-09-21 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures at 2.2-A resolution of hydroxyethylene-based inhibitors bound to human immunodeficiency virus type 1 protease show that the inhibitors are present in two distinct orientations.
J.Biol.Chem., 267, 1992
|
|
1HEH
 
 | | C-terminal xylan binding domain from Cellulomonas fimi xylanase 11A | | Descriptor: | ENDO-1,4-BETA-XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, White, P, Hancock, S.M, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Evidence for Synergy between Family 2B Carbohydrate Binding Modules in Cellulomonas Fimi Xylanase 11A
Biochemistry, 40, 2001
|
|
1HEI
 
 | |
1HEJ
 
 | | C-terminal xylan binding domain from Cellulomonas fimi xylanase 11A | | Descriptor: | ENDO-1,4-BETA-XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, White, P, Hancock, S.M, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Evidence for Synergy between Family 2B Carbohydrate Binding Modules in Cellulomonas Fimi Xylanase 11A
Biochemistry, 40, 2001
|
|
1HEK
 
 | | Crystal structure of equine infectious anaemia virus matrix antigen (EIAV MA) | | Descriptor: | GAG POLYPROTEIN, CORE PROTEIN P15 | | Authors: | Hatanaka, H, Iourin, O, Rao, Z, Fry, E, Kingsman, A, Stuart, D.I. | | Deposit date: | 2000-11-24 | | Release date: | 2001-11-23 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Equine Infectious Anemia Virus Matrix Protein.
J.Virol., 76, 2002
|
|
1HEL
 
 | |
1HEM
 
 | |
1HEN
 
 | |
1HEO
 
 | |
1HEP
 
 | |
1HEQ
 
 | |
1HER
 
 | |
1HES
 
 | |
1HET
 
 | | atomic X-ray structure of liver alcohol dehydrogenase containing a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-25 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1HEU
 
 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1HEV
 
 | |
1HEW
 
 | |
1HEX
 
 | |
1HEY
 
 | |
1HEZ
 
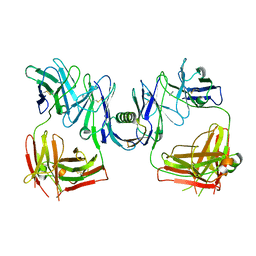 | | Structure of P. magnus protein L bound to a human IgM Fab. | | Descriptor: | HEAVY CHAIN OF IG, IMIDAZOLE, KAPPA LIGHT CHAIN OF IG, ... | | Authors: | Graille, M, Stura, E.A, Housden, N.G, Bottomley, S.P, Taussig, M.J, Sutton, B.J, Gore, M.G, Charbonnier, J.B. | | Deposit date: | 2000-11-27 | | Release date: | 2001-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex between Peptostreptococcus Magnus Protein L and a Human Antibody Reveals Structural Convergence in the Interaction Modes of Fab Binding Modes
Structure, 9, 2001
|
|
1HF0
 
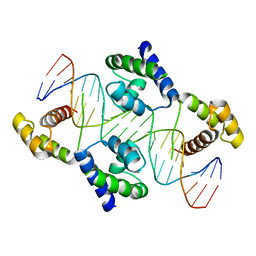 | | Crystal structure of the DNA-binding domain of Oct-1 bound to DNA as a dimer | | Descriptor: | DNA 5'-D(*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP* CP*AP*AP*AP*TP*GP*GP*AP*G)-3', DNA 5'-D(*CP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP* TP*CP*AP*AP*AP*TP*GP*TP*G)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | Authors: | Remenyi, A, Tomilin, A, Pohl, E, Scholer, H.R, Wilmanns, M. | | Deposit date: | 2000-11-27 | | Release date: | 2001-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
1HF2
 
 | |
1HF3
 
 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-27 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1HF4
 
 | | STRUCTURAL EFFECTS OF MONOVALENT ANIONS ON POLYMORPHIC LYSOZYME CRYSTALS | | Descriptor: | LYSOZYME, NITRATE ION, SODIUM ION | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 2000-11-29 | | Release date: | 2001-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Effects of Monovalent Anions on Polymorphic Lysozyme Crystals
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HF6
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE | | Descriptor: | ACETIC ACID, ENDOGLUCANASE B, GLYCEROL, ... | | Authors: | Varrot, A, Withers, S, Vasella, A, Schulein, M, Davies, G.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct Experimental Observation of the Hydrogen-Bonding Network of a Glycosidase Along its Reaction Coordinate Revealed by Atomic Resolution Analyses of Endoglucanase Cel5A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
