7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
6YQ4
 
 | | Crystal structure of Fusobacterium nucleatum tannase | | Descriptor: | GLYCEROL, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Mancheno, J.M, Anguita, J, Rodriguez, H. | | Deposit date: | 2020-04-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | A structurally unique Fusobacterium nucleatum tannase provides detoxicant activity against gallotannins and pathogen resistance.
Microb Biotechnol, 15, 2022
|
|
8BA6
 
 | | Structure of the FK1 domain of the FKBP51 G64S variant in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{R})-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Hausch, F. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Binding pocket stabilization by high-throughput screening of yeast display libraries.
Front Mol Biosci, 9, 2022
|
|
8BAJ
 
 | | Structure of the FK1 domain of the FKBP51 G64S variant in complex with (1S,5S,6R)-10-((3,5-dichlorophenyl)sulfonyl)-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Hausch, F. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Binding pocket stabilization by high-throughput screening of yeast display libraries.
Front Mol Biosci, 9, 2022
|
|
6W5K
 
 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5g | | Descriptor: | 3C-LIKE PROTEASE, N~2~-{[2-(3-chlorophenyl)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
3QTE
 
 | |
7ZJZ
 
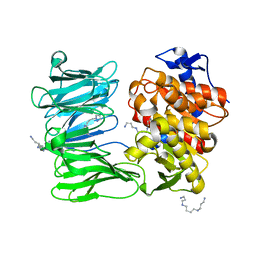 | | catalytically non active S532A mutant of oligopeptidase B from S. proteomaculans | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-04-12 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
6W5H
 
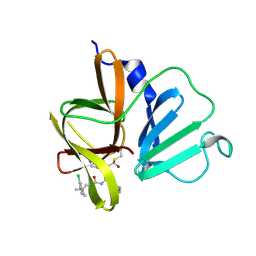 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5d | | Descriptor: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6W6D
 
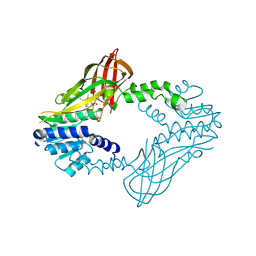 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with SGC6870 inhibitor | | Descriptor: | (5R)-4-(5-bromothiophene-2-carbonyl)-5-(3,5-dimethylphenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Jin, J, Shen, Y, Kaniskan, H.U, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A First-in-Class, Highly Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 6.
J.Med.Chem., 64, 2021
|
|
6W78
 
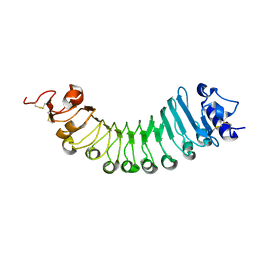 | | crystal structure of a plant ice-binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antifreeze polypeptide | | Authors: | Wang, Y.N, Zhang, H.Q. | | Deposit date: | 2020-03-18 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Carrot 'antifreeze' protein has an irregular ice-binding site that confers weak freezing point depression but strong inhibition of ice recrystallization.
Biochem.J., 477, 2020
|
|
3Q2V
 
 | | Crystal structure of mouse E-cadherin ectodomain | | Descriptor: | CALCIUM ION, Cadherin-1, MANGANESE (II) ION, ... | | Authors: | Jin, X, Harrison, O.J, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
6YMN
 
 | |
6VYO
 
 | | Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
3Q2W
 
 | | Crystal structure of mouse N-cadherin ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
6W5L
 
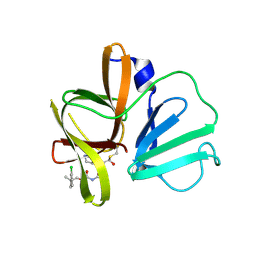 | | 2.1 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7g | | Descriptor: | (2~{S})-~{N}-[(1~{R})-1-[bis($l^{1}-oxidanyl)-methoxy-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[[2-(3-chlorophenyl)-2-methyl-propoxy]-oxidanylidene-methyl]amino]-4-methyl-pentanamide, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6VWC
 
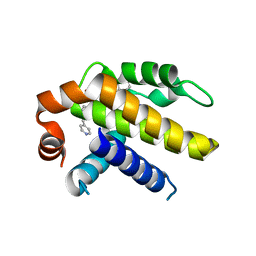 | | Crystal structure of Bcl-xL in complex with tetrahydroisoquinoline-pyridine based inhibitors | | Descriptor: | 6-{8-[(1,3-benzothiazol-2-yl)carbamoyl]-3,4-dihydroisoquinolin-2(1H)-yl}-3-{1-[(pyridin-4-yl)methyl]-1H-pyrazol-4-yl}pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Judge, R.A, Judd, A.S. | | Deposit date: | 2020-02-19 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Discovery of A-1331852, a First-in-Class, Potent, and Orally-Bioavailable BCL-X L Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
6VWF
 
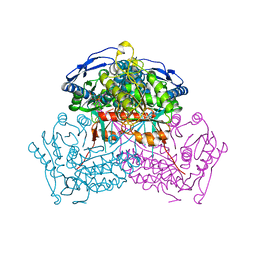 | | Structure of ALDH9A1 complexed with NAD+ in space group C222 | | Descriptor: | 4-trimethylaminobutyraldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wyatt, J.W, Tanner, J.J. | | Deposit date: | 2020-02-19 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch.Biochem.Biophys., 691, 2020
|
|
6WOK
 
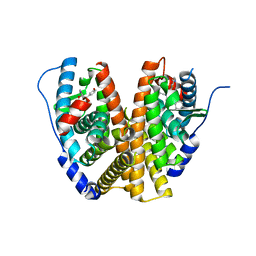 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 6 | | Descriptor: | (1R,3R)-1-(2,6-difluoro-4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Labadie, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Discovery of GNE-149 as a Full Antagonist and Efficient Degrader of Estrogen Receptor alpha for ER+ Breast Cancer.
Acs Med.Chem.Lett., 11, 2020
|
|
8BA4
 
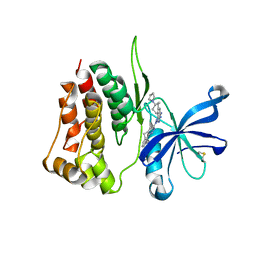 | | Crystal structure of JAK2 JH2-V617F in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BA3
 
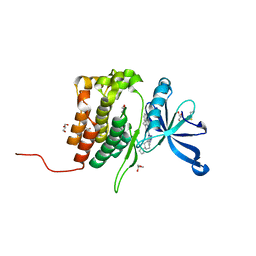 | | Crystal structure of JAK2 JH2 in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8U
 
 | |
7ZGV
 
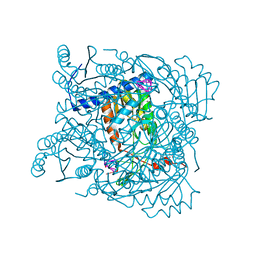 | | Serratia NucC bound to cA3 | | Descriptor: | ACETATE ION, CALCIUM ION, RNA (5'-R(P*AP*AP*A)-3'), ... | | Authors: | Garcia-Doval, C, Mayo-Munoz, D, Smith, L.M, Fineran, P.C. | | Deposit date: | 2022-04-04 | | Release date: | 2022-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Type III CRISPR-Cas provides resistance against nucleus-forming jumbo phages via abortive infection.
Mol.Cell, 82, 2022
|
|
7ZRA
 
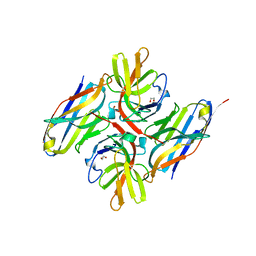 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS1(Nb14497) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody NbSOS1 (Nb14497) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2022-05-04 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
7ZGW
 
 | |
3RS6
 
 | | Crystal structure Dioclea virgata lectin in complexed with X-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Lectin alpha chain, ... | | Authors: | Gadelha, C.A.A, Santi-Gadelha, T, Nagano, C.S, Bezerra, E.H.S, Bezerra, M.J.B, Alencar, K.L, Silva-Filho, J.C. | | Deposit date: | 2011-05-02 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Dioclea virgata lectin: Relations between carbohydrate binding site and nitric oxide production.
Biochimie, 94, 2012
|
|
