7JMJ
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 37 - State 5 (S5) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
7JMI
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 29 - State 3 (S3) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
7JMG
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 22 - State 2 (S2) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
7JMF
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 42 - State 6 (S6) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
3EJV
 
 | |
3EBY
 
 | |
3ES1
 
 | |
4P3X
 
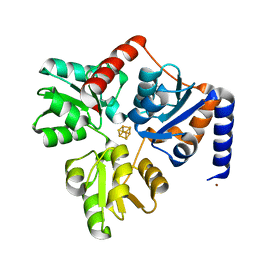 | | Structure of the Fe4S4 quinolinate synthase NadA from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Cherrier, M.V, Chan, A, Darnault, C, Reichmann, D, Amara, P, Ollagnier de Choudens, S, Fontecilla-Camps, J.C. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of Fe4S4 quinolinate synthase unravels an enzymatic dehydration mechanism that uses tyrosine and a hydrolase-type triad.
J.Am.Chem.Soc., 136, 2014
|
|
1R76
 
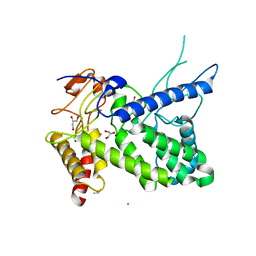 | | Structure of a pectate lyase from Azospirillum irakense | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Novoa de Armas, H, Verboven, C, De Ranter, C, Desair, J, Vande Broek, A, Vanderleyden, J, Rabijns, A. | | Deposit date: | 2003-10-20 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Azospirillum irakense pectate lyase displays a toroidal fold.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6PV6
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2019-07-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
2RAS
 
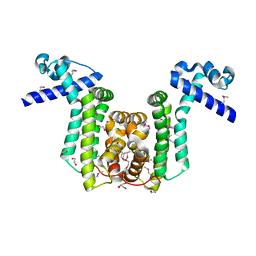 | |
3B8L
 
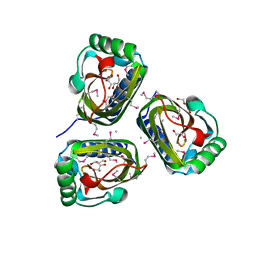 | |
3BWX
 
 | |
3M7A
 
 | |
3CJY
 
 | |
3FF2
 
 | |
3G0K
 
 | |
3EC4
 
 | |
2QTQ
 
 | |
3C5Y
 
 | |
4K8G
 
 | | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans mutant (V161A, R163A, K165G, L166A, Y167G, Y168A, E169G) | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Lukk, T, Wichelecki, D, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans mutant (V161A, R163A, K165G, L166A, Y167G, Y168A, E169G)
To be Published
|
|
6R0V
 
 | |
6R0U
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 5a and hydrolysis product | | Descriptor: | 3-azanyl-2-[[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]carbamoyl]benzoic acid, 4-azanyl-2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]isoindole-1,3-dione, CHLORIDE ION, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
6R0S
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 4a and hydrolysis product | | Descriptor: | 2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]-4-nitro-isoindole-1,3-dione, 2-[[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]carbamoyl]-6-nitro-benzoic acid, CEREBLON ISOFORM 4, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
6R0Q
 
 | |
