6RNK
 
 | | Crystal structure of a humanized (K18E, K269N) rat succinate receptor SUCNR1 (GPR91) in complex with a nanobody and antagonist NF-56-EJ40. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[2-[[3-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, GLYCEROL, ... | | Authors: | Haffke, M, Jaakola, V.-P. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis of species-selective antagonist binding to the succinate receptor.
Nature, 574, 2019
|
|
4ZZN
 
 | | Human ERK2 in complex with an inhibitor | | Descriptor: | 2-[[5-chloranyl-2-(oxan-4-ylamino)pyridin-4-yl]amino]-N-methyl-benzamide, MITOGEN-ACTIVATED PROTEIN KINASE 1, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
9F4O
 
 | | UP1 in complex with Z991506900 | | Descriptor: | (3~{R})-3-methyl-1-(6-methylpyridin-2-yl)piperazine, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
6RFO
 
 | | ERK2 MAP kinase with the activation loop of p38alpha | | Descriptor: | Mitogen-activated protein kinase 1,Mitogen-activated protein kinase 14,Mitogen-activated protein kinase 1 | | Authors: | Livnah, O, Eitan-Wexler, M. | | Deposit date: | 2019-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The bacterial metalloprotease NleD selectively cleaves mitogen-activated protein kinases that have high flexibility in their activation loop.
J.Biol.Chem., 295, 2020
|
|
6RO9
 
 | | Human spectrin SH3 domain D48G, E7V, K60V | | Descriptor: | Spectrin alpha, non-erythrocytic 1 | | Authors: | Reverter, D, Navarro, S, Ventura, S. | | Deposit date: | 2019-05-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Human spectrin SH3 domain D48G, E7V, K60V
To Be Published
|
|
6ROA
 
 | | Crystal structure of V57G mutant of human cystatin C | | Descriptor: | Cystatin-C | | Authors: | Orlikowska, M, Behrendt, I, Borek, D, Otwinowski, Z, Skowron, P, Szymanska, A. | | Deposit date: | 2019-05-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | NMR and crystallographic structural studies of the extremely stable monomeric variant of human cystatin C with single amino acid substitution.
Febs J., 287, 2020
|
|
9EO2
 
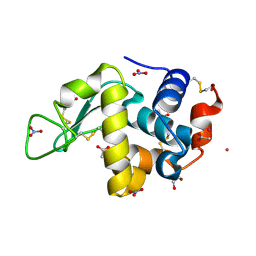 | |
3KX4
 
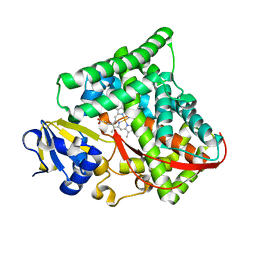 | | Crystal structure of Bacillus megaterium BM3 heme domain mutant I401E | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Girvan, H.M, Levy, C.W, Leys, D, Munro, A.W. | | Deposit date: | 2009-12-02 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Glutamate-haem ester bond formation is disfavoured in flavocytochrome P450 BM3: characterization of glutamate substitution mutants at the haem site of P450 BM3.
Biochem.J., 427, 2010
|
|
5A5I
 
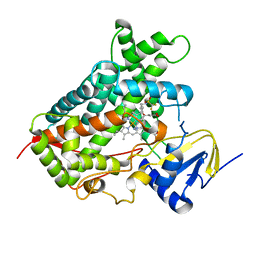 | | Cytochrome 2C9 P450 inhibitor complex | | Descriptor: | CYTOCHROME P450 2C9, N-[4-(3-chloranyl-4-cyano-phenoxy)cyclohexyl]-1,1,1-tris(fluoranyl)methanesulfonamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Skerratt, S.E, de Groot, M.J, Phillips, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Novel Binding Pocket for Cyp 2C9 Inhibitors: Crystallography, Pharmacophore Modelling and Inhibitor Sar.
Med.Chem.Comm., 7, 2016
|
|
9ENZ
 
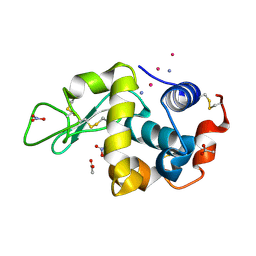 | |
8ZJV
 
 | | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Yang, Y, Fu, L, Chen, R, Cheng, H, Sun, X. | | Deposit date: | 2024-05-15 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin
To Be Published
|
|
9F52
 
 | | UP1 in complex with Z734147462 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, [4-(methylamino)oxan-4-yl]methanol | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4K
 
 | | UP1 in complex with ZINC72259689 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, ~{N}-(cyclobutylmethyl)-1,3-dimethyl-pyrazole-4-carboxamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
4ZSQ
 
 | | BACE crystal structure with tricyclic aminothiazine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(4S,4aS,6S,8aR)-10-aminohexahydro-3H-4,8a-(epithiomethenoazeno)isochromen-6(1H)-yl]-3-chlorobenzamide | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
9F55
 
 | | UP1 in complex with Z235361315 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, ~{N}-(3-azanyl-2-methyl-phenyl)propanamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
4ZTQ
 
 | | Human Aurora A catalytic domain bound to FK932 | | Descriptor: | (2Z,5Z)-2-[(4-ethylphenyl)imino]-3-(2-methoxyethyl)-5-(pyridin-4-ylmethylidene)-1,3-thiazolidin-4-one, (4S)-2-METHYL-2,4-PENTANEDIOL, Aurora kinase A | | Authors: | Marcaida, M.J, Kilchmann, F, Schick, T, Reymond, J.L. | | Deposit date: | 2015-05-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Selective Aurora A Kinase Inhibitor by Virtual Screening.
J.Med.Chem., 59, 2016
|
|
6RSK
 
 | |
9EO8
 
 | |
6RSV
 
 | | Endothiapepsin in complex with 017 | | Descriptor: | 1~{H}-1,2,3,4-tetrazol-5-ylmethyldiazane, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Magari, F, Heine, A, Klebe, G. | | Deposit date: | 2019-05-22 | | Release date: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Endothiapepsin in complex with 017
To Be Published
|
|
3L3C
 
 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to Glc6P | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-12-16 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3L4I
 
 | | CRYSTAL STRUCTURE OF THE N-TERMINAL DOMAIN OF HSP70 (CGD2_20) FROM Cryptosporidium PARVUM IN COMPLEX WITH ADP and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Sullivan, H, Lew, J, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-20 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE N-TERMINAL DOMAIN OF HSP70 (CGD2_20) FROM Cryptosporidium PARVUM IN COMPLEX WITH ADP and inorganic phosphate
To be Published
|
|
3KL8
 
 | | CaMKIINtide Inhibitor Complex | | Descriptor: | Calcium/calmodulin dependent protein kinase II, Calcium/calmodulin-dependent protein kinase II inhibitor 1 | | Authors: | Kuriyan, J, Chao, L.H, Pellicena, P, Deindl, S, Barclay, L.A, Schulman, H. | | Deposit date: | 2009-11-06 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.372 Å) | | Cite: | Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5AEQ
 
 | | Neuronal calcium sensor (NCS-1)from Rattus norvegicus | | Descriptor: | CALCIUM ION, NEURONAL CALCIUM SENSOR 1, SODIUM ION | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
9EO5
 
 | |
3KKO
 
 | | Crystal structure of M-Ras P40D/D41E/L51R in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
