6DKD
 
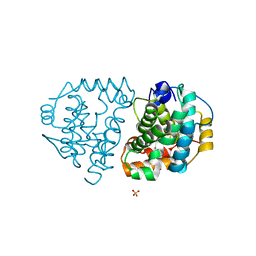 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
6DKC
 
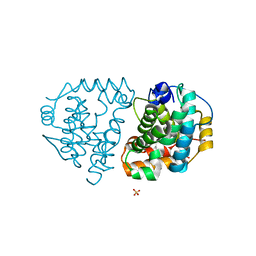 | | Yeast Ddi2 Cyanamide Hydratase, T157V mutant, apo structure | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
9JKD
 
 | |
9JKC
 
 | |
4P8H
 
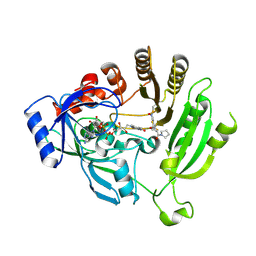 | | Crystal structure of M. tuberculosis DprE1 in complex with the nitro-benzothiazole 6a | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable decaprenylphosphoryl-beta-D-ribose oxidase, {(4S)-2-[7-(hydroxyamino)-6-methyl-5-(trifluoromethyl)-1,3-benzothiazol-2-yl]-4,5-dihydro-1,3-oxazol-4-yl}(pyrrolidin-1-yl)methanone | | Authors: | Neres, J, Cole, S.T. | | Deposit date: | 2014-03-31 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Non-mutagenic Nitrobenzothiazoles as Novel Anti-tubercular Agents: A Balance between Potency and Electron Affinity
To Be Published
|
|
3CXY
 
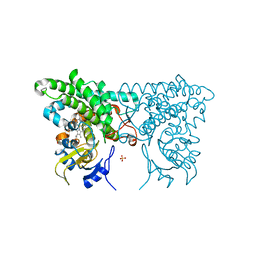 | |
3CM1
 
 | |
9FIB
 
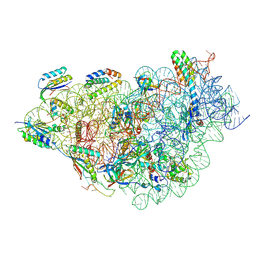 | | Structure of 30S-IF1-IF3-mRNA-GE81112A complex | | Descriptor: | (2S,3S)-2-[[(2S)-2-[[(2S,4S)-5-aminocarbonyloxy-4-oxidanyl-2-[[(2S,3R)-3-oxidanylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-3-(1H-imidazol-4-yl)propanoyl]amino]-3-(2-chloranyl-1H-imidazol-4-yl)-3-oxidanyl-propanoic acid, 16S rRNA, MAGNESIUM ION, ... | | Authors: | Safdari, H.A, Morici, M, Wilson, D.N. | | Deposit date: | 2024-05-29 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | The translation inhibitors kasugamycin, edeine and GE81112 target distinct steps during 30S initiation complex formation.
Nat Commun, 16, 2025
|
|
9FDA
 
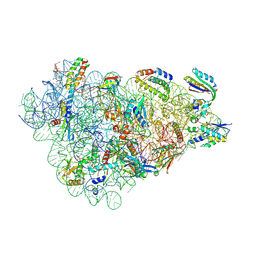 | |
3HO2
 
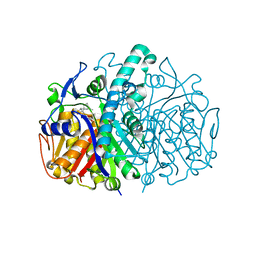 | | Structure of E.coli FabF(C163A) in complex with Platencin | | Descriptor: | 2,4-dihydroxy-3-({3-[(2S,4aS,8S,8aR)-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonaphthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-06-01 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isolation, enzyme-bound structure and antibacterial activity of platencin A1 from Streptomyces platensis.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
9G06
 
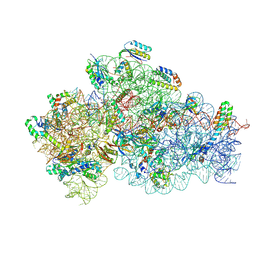 | | Structure of 30S-IF1-IF3-mRNA-fMet-tRNA-GE81112A complex | | Descriptor: | (2S,3S)-2-[[(2S)-2-[[(2S,4S)-5-aminocarbonyloxy-4-oxidanyl-2-[[(2S,3R)-3-oxidanylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-3-(1H-imidazol-4-yl)propanoyl]amino]-3-(2-chloranyl-1H-imidazol-4-yl)-3-oxidanyl-propanoic acid, 16S ribosomal RNA, MAGNESIUM ION, ... | | Authors: | Safdari, H.A, Morici, M, Wilson, D.N. | | Deposit date: | 2024-07-07 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | The translation inhibitors kasugamycin, edeine and GE81112 target distinct steps during 30S initiation complex formation.
Nat Commun, 16, 2025
|
|
8CAS
 
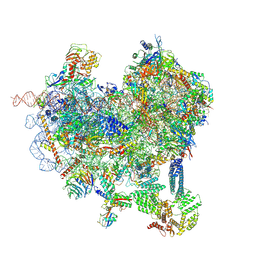 | | Cryo-EM structure of native Otu2-bound ubiquitinated 48S initiation complex (partial) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8CBJ
 
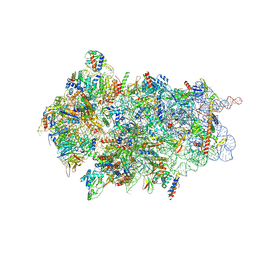 | | Cryo-EM structure of Otu2-bound cytoplasmic pre-40S ribosome biogenesis complex | | Descriptor: | 20S pre-ribosomal RNA, 20S-pre-rRNA D-site endonuclease NOB1, 40S ribosomal protein S0-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8CLT
 
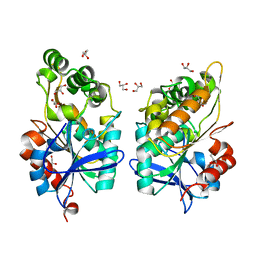 | |
8CLU
 
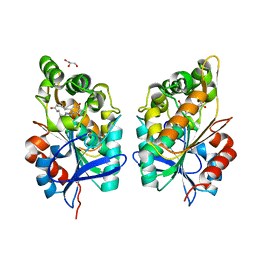 | | Zearalenone lactonase from Rhodococcus erythropolis in complex with zearalactamenone | | Descriptor: | (4~{S})-4-methyl-16,18-bis(oxidanyl)-3-azabicyclo[12.4.0]octadeca-1(18),12,14,16-tetraene-2,8-dione, GLYCEROL, Zearalenone lactonase | | Authors: | Puehringer, D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
3HO9
 
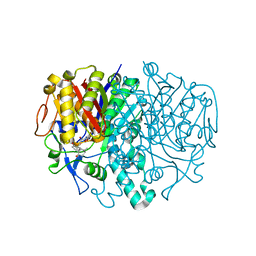 | | Structure of E.coli FabF(C163A) in complex with Platencin A1 | | Descriptor: | 2,4-dihydroxy-3-({3-[(2R,4aR,8S,8aR,9R)-9-hydroxy-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonap hthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-06-01 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isolation, enzyme-bound structure and antibacterial activity of platencin A1 from Streptomyces platensis.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1X8V
 
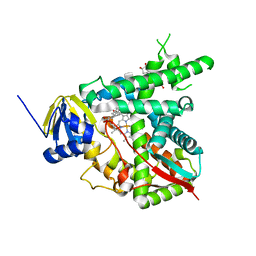 | | Estriol-bound and ligand-free structures of sterol 14alpha-demethylase (CYP51) | | Descriptor: | Cytochrome P450 51, ESTRIOL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M, Yermalitskaya, L.V, Lepesheva, G.I, Podust, V.N, Dalmasso, E.A, Waterman, M.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Estriol Bound and Ligand-free Structures of Sterol 14alpha-Demethylase.
Structure, 12, 2004
|
|
4KZW
 
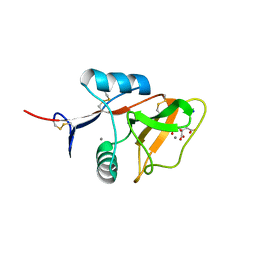 | | Structure of the carbohydrate-recognition domain of the C-type lectin mincle | | Descriptor: | C-TYPE LECTIN MINCLE, CALCIUM ION, CITRATE ANION, ... | | Authors: | Feinberg, H, Jegouzo, S.A.F, Rowntree, T.J.W, Guan, Y, Brash, M.A, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2013-05-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism for Recognition of an Unusual Mycobacterial Glycolipid by the Macrophage Receptor Mincle.
J.Biol.Chem., 288, 2013
|
|
1TDT
 
 | |
4KZV
 
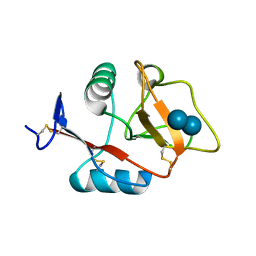 | | Structure of the carbohydrate-recognition domain of the C-type lectin mincle bound to trehalose | | Descriptor: | C-type lectin mincle, CALCIUM ION, SODIUM ION, ... | | Authors: | Feinberg, H, Jegouzo, S.A.F, Rowntree, T.J.W, Guan, Y, Brash, M.A, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2013-05-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism for Recognition of an Unusual Mycobacterial Glycolipid by the Macrophage Receptor Mincle.
J.Biol.Chem., 288, 2013
|
|
3C0E
 
 | |
8ZPK
 
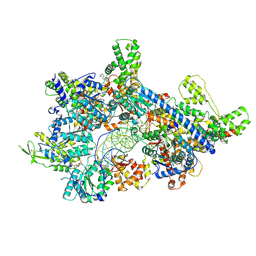 | | Cryo-EM structure of origin recognition complex (Orc6 with residues 1 to 270 deleted) with ARS1 DNA bound | | Descriptor: | DNA (38-MER), DNA (40-MER), MAGNESIUM ION, ... | | Authors: | Lam, W.H, Yu, D, Dang, S, Zhai, Y. | | Deposit date: | 2024-05-30 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | DNA bending mediated by ORC is essential for replication licensing in budding yeast.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8ZP5
 
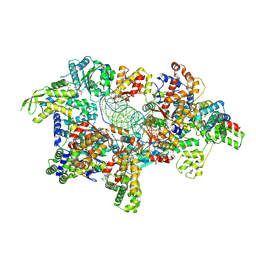 | | Cryo-EM structure of origin recognition complex (Orc5 basic patch mutations) with ARS1 DNA bound | | Descriptor: | DNA (34-MER), DNA (35-MER), MAGNESIUM ION, ... | | Authors: | Lam, W.H, Yu, D, Dang, S, Zhai, Y. | | Deposit date: | 2024-05-29 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | DNA bending mediated by ORC is essential for replication licensing in budding yeast.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
4Y59
 
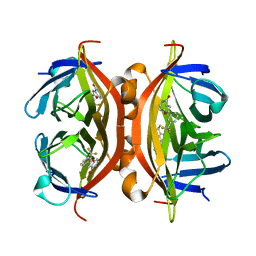 | | Crystal structure of ALiS1-Streptavidin complex | | Descriptor: | 2-[3-(trifluoromethyl)phenyl]furo[3,2-c]pyridin-4(5H)-one, Streptavidin | | Authors: | Sugiyama, S, Terai, T, Kohno, M, Ishida, H, Nagano, T. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Artificial Ligands of Streptavidin (ALiS): Discovery, Characterization, and Application for Reversible Control of Intracellular Protein Transport
J.Am.Chem.Soc., 137, 2015
|
|
4Y5D
 
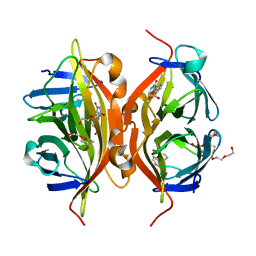 | | CRYSTAL STRUCTURE OF ALiS2-STREPTAVIDIN COMPLEX | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, DIMETHYL SULFOXIDE, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Terai, T, Kohno, M, Ishida, H, Nagano, T. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Artificial Ligands of Streptavidin (ALiS): Discovery, Characterization, and Application for Reversible Control of Intracellular Protein Transport
J.Am.Chem.Soc., 137, 2015
|
|
