8RNG
 
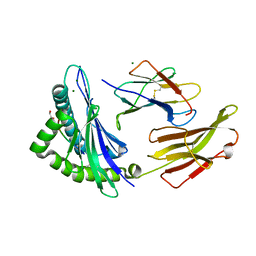 | | Crystal structure of HLA B*18:01 in complex with TEVETYVL, an 8-mer epitope from Influenza A | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MAGNESIUM ION, ... | | Authors: | Murdolo, L.D, Maddumage, J.C, Gras, S. | | Deposit date: | 2024-01-10 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of HLA B*18:01 in complex with TEVETYVL, an 8-mer epitope from Influenza A
To Be Published
|
|
5D9R
 
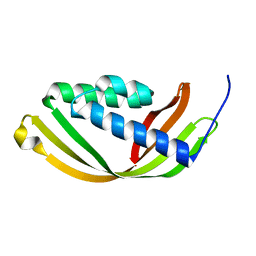 | | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6 | | Descriptor: | Protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic | | Authors: | Radhakrishnan, A, Kumar, N, Su, C.-C, Chou, T.-H, Yu, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6.
Protein Sci., 25, 2016
|
|
8RNH
 
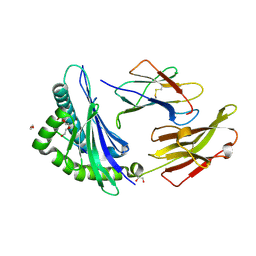 | | Crystal structure of HLA B*18:01 in complex with EEIEITTHF, an 9-mer epitope from Influenza A | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murdolo, L.D, Maddumaage, J, Gras, S. | | Deposit date: | 2024-01-10 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HLA B*18:01 in complex with EEIEITTHF, an 9-mer epitope from Influenza A
To Be Published
|
|
8SPA
 
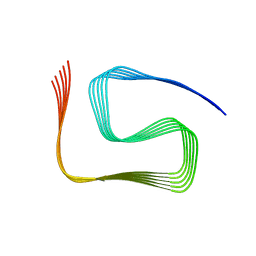 | | Structural insights into cellular control of the human CPEB3 prion, functionally regulated by a labile-amyloid-forming segment | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Flores, M.D, Sawaya, M.R, Boyer, D.R, Zink, S, Fioriti, L, Rodriguez, J.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a reversible amyloid fibril formed by the CPEB3 prion-like domain reveals a core sequence involved in translational regulation
To Be Published
|
|
8SG2
 
 | | BIVALENT INTERACTIONS OF PIN1 WITH THE C-TERMINAL TAIL OF PKC | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, Protein kinase C beta type | | Authors: | Dixit, K, Yang, Y, Chen, X.R, Igumenova, T.I. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel bivalent interaction mode underlies a non-catalytic mechanism for Pin1-mediated protein kinase C regulation.
Elife, 13, 2024
|
|
8RFK
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - single mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, Quinoprotein glucose dehydrogenase B | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8SGK
 
 | | CryoEM structure of Deinococcus radiodurans BphP photosensory module in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, H, Li, H. | | Deposit date: | 2023-04-12 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CryoEM structure of Deinococcus radiodurans BphP photosensory module in Pr state
To Be Published
|
|
5D60
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form III | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D69
 
 | | Human calpain PEF(S) with (2Z,2Z')-2,2'-disulfanediylbis(3-(6-iodoindol-3-yl)acrylic acid) bound | | Descriptor: | (2E,2'Z)-2,2'-disulfanediylbis[3-(4-iodophenyl)prop-2-enoic acid], CALCIUM ION, Calpain small subunit 1, ... | | Authors: | Adams, S.E, Robinson, E.J, Rizkallah, P.J, Miller, D.J, Hallett, M.B, Allemann, R.K. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Conformationally restricted calpain inhibitors.
Chem Sci, 6, 2015
|
|
8R9U
 
 | | A soakable crystal form of human CDK7 in complex with AMP-PNP | | Descriptor: | 7-dimethylphosphoryl-3-[2-[[(3~{S})-6,6-dimethylpiperidin-3-yl]amino]-5-(trifluoromethyl)pyrimidin-4-yl]-1~{H}-indole-6-carbonitrile, Cyclin-dependent kinase 7 | | Authors: | Mukherjee, M, Cleasby, A. | | Deposit date: | 2023-11-30 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Protein engineering enables a soakable crystal form of human CDK7 primed for high-throughput crystallography and structure-based drug design.
Structure, 2024
|
|
2FQ5
 
 | |
2FW9
 
 | |
5D6X
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
8RE0
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - double mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8R67
 
 | | tubulin-cryptophycin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(3~{S},10~{R},13~{E},16~{S})-10-[(3-chloranyl-4-methoxy-phenyl)methyl]-6,6-dimethyl-2,5,9,12-tetrakis(oxidanylidene)-16-[(1~{S})-1-[(2~{R},3~{R})-3-phenyloxiran-2-yl]ethyl]-1,4-dioxa-8,11-diazacyclohexadec-13-en-3-yl]ethanoic acid, CALCIUM ION, ... | | Authors: | Abel, A.C, Muehlethaler, T, Dessin, C, Steinmetz, M.O, Sewald, N, Prota, A.E. | | Deposit date: | 2023-11-21 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bridging the maytansine and vinca sites: Cryptophycins target beta-tubulin's T5-loop.
J.Biol.Chem., 300, 2024
|
|
8RZB
 
 | | IL-1beta in complex with covalent DEL hit | | Descriptor: | 8-[4-methyl-3-(trifluoromethyl)phenyl]-2-[[(7S)-7-(2-morpholin-4-ylethylcarbamoyl)-4-(phenylsulfonyl)-1,4-diazepan-1-yl]carbonyl]imidazo[1,2-a]pyridine-6-carboxylic acid, Interleukin-1 beta | | Authors: | Rondeau, J.-M, Lehmann, S. | | Deposit date: | 2024-02-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Ligandability Assessment of IL-1 beta by Integrated Hit Identification Approaches.
J.Med.Chem., 67, 2024
|
|
5D7C
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[1-(pyridin-2-yl)-6-(pyridin-3-yl)-1H-pyrrolo[3,2-b]pyridin-3-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5D7R
 
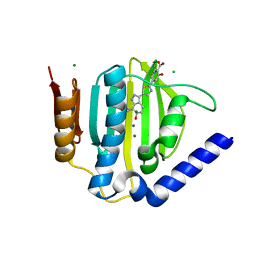 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-hydroxy-5-[5-(6-hydroxy-7-propyl-2H-indazol-3-yl)-1,3-thiazol-2-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-14 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
8RFM
 
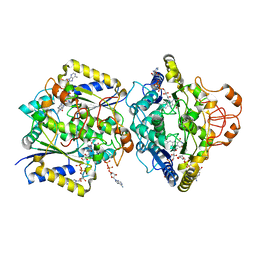 | | Human NOQ1 enzyme in complex with NADH by serial crystallography | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Martin-Garcia, J.M, Grieco, A, Medina, M, Boneta, S, Pey, A.L. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural dynamics and functional cooperativity of human NQO1 by ambient temperature serial crystallography and simulations.
Protein Sci., 33, 2024
|
|
8SNL
 
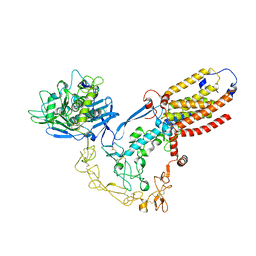 | | Structure of human ADAM17/iRhom2 sheddase complex | | Descriptor: | CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 17, Inactive rhomboid protein 2, ... | | Authors: | Zhao, H, Dai, Y, Wang, Y, Lee, C.H. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structure of human ADAM17/iRhom2 sheddase complex
To Be Published
|
|
2FXR
 
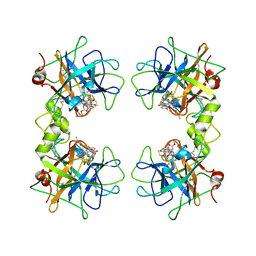 | | human beta tryptase II complexed with activated ketone inhibitor CRA-29382 | | Descriptor: | ALLYL {(1S)-1-[(5-{4-[(2,3-DIHYDRO-1H-INDEN-2-YLAMINO)CARBONYL]BENZYL}-1,2,4-OXADIAZOL-3-YL)CARBONYL]-3-PYRROLIDIN-3-YLPROPYL}CARBAMATE, Tryptase beta-2 | | Authors: | Katz, B.A. | | Deposit date: | 2006-02-06 | | Release date: | 2006-02-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
5D8D
 
 | |
8RCG
 
 | | W-formate dehydrogenase M405S from Desulfovibrio vulgaris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural and biochemical characterization of the M405S variant of Desulfovibrio vulgaris formate dehydrogenase.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
3UV7
 
 | | Ec_IspH in complex with buta-2,3-dienyl diphosphate (1300) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, buta-2,3-dien-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
2FYT
 
 | | Human HMT1 hnRNP methyltransferase-like 3 (S. cerevisiae) protein | | Descriptor: | Protein arginine N-methyltransferase 3, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-08 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human HMT1 hnRNP
methyltransferase-like 3 in complex with SAH.
To be Published
|
|
