4EPE
 
 | | Final Urease Structure for Radiation Damage Experiment at 300 K | | Descriptor: | NICKEL (II) ION, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | Deposit date: | 2012-04-17 | | Release date: | 2012-08-29 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SR1
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS A69G bound to Ca2+ and thymidine-5',3'-diphosphate at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, thermonuclease | | Authors: | Doctrow, B.M, Schlessman, J.L, Garcia-Moreno E, B, Heroux, A. | | Deposit date: | 2011-07-06 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS A69G bound to Ca2+ and thymidine-5',3'-diphosphate at cryogenic temperature
To be Published
|
|
3E19
 
 | | Crystal Structure of Iron Uptake Regulatory Protein (FeoA) Solved by Sulfur SAD in a Monoclinic Space Group | | Descriptor: | FeoA, GLYCEROL, PHOSPHATE ION | | Authors: | Hughes, R.C, Li, Y, Wang, B.-C, Liu, Z.-J, Ng, J.D. | | Deposit date: | 2008-08-02 | | Release date: | 2008-12-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Structure Determination of Iron Uptake Regulatory
Protein (FeoA) by Sulfur SAD in a Monoclinic Space Group
To be Published
|
|
3DYI
 
 | |
3SRU
 
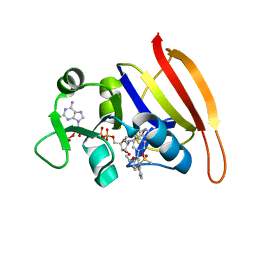 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | Dihydrofolate reductase, N-[3'-(2,4-diaminoquinazolin-7-yl)-4'-ethoxybiphenyl-3-yl]methanesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4EKS
 
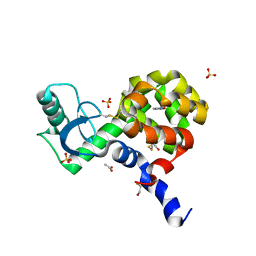 | | T4 Lysozyme L99A/M102H with Isoxazole Bound | | Descriptor: | 1,2-benzisoxazole, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Engineering a model protein cavity to catalyze the Kemp elimination.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FEA
 
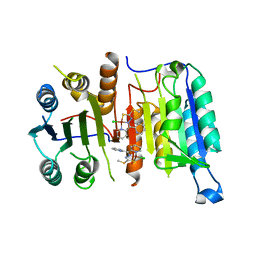 | | Crystal structure of CASPASE-7 in Complex with allosteric inhibitor | | Descriptor: | Caspase-7, chloro{methyl hydrogenato(3-)-kappa~2~N,S [pyridin-2-yl(pyridin-2(1H)-ylidene-kappaN)methyl]carbonodithiohydrazonate}copper | | Authors: | Kabaleeswaran, V. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | A class of allosteric caspase inhibitors identified by high-throughput screening.
Mol.Cell, 47, 2012
|
|
4EL7
 
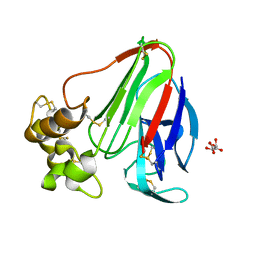 | | Initial Thaumatin Structure for Radiation Damage Experiment at 300 K | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-29 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3DV6
 
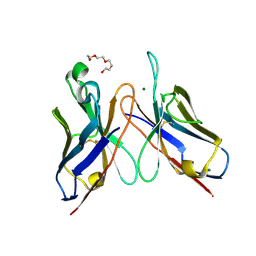 | | Crystal structure of SAG506-01, tetragonal, crystal 2 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-18 | | Release date: | 2008-12-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3STC
 
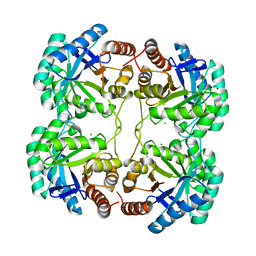 | |
3DZO
 
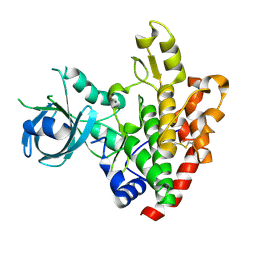 | | Crystal structure of a rhoptry kinase from toxoplasma gondii | | Descriptor: | MAGNESIUM ION, Rhoptry kinase domain | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Ni, S, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Sibley, D, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
4ENT
 
 | | Structure of the S234A variant of E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-02 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of main channel structure on H(2)O(2) access to the heme cavity of catalase KatE of Escherichia coli.
Arch.Biochem.Biophys., 526, 2012
|
|
3DX7
 
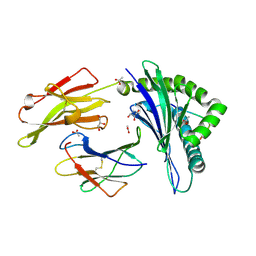 | | Crystal Structure of HLA-B*4403 presenting 10mer EBV antigen | | Descriptor: | ACETATE ION, Beta-2-microglobulin, EBV decapeptide epitope, ... | | Authors: | Archbold, J.K, Ely, L.K, Rossjohn, J. | | Deposit date: | 2008-07-23 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition.
J.Exp.Med., 206, 2009
|
|
4GGQ
 
 | |
4EQJ
 
 | | Crystal Structure of inactive single chain variant of HIV-1 Protease in Complex with the substrate RT-RH | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-19 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
3E1W
 
 | |
3DXN
 
 | | Crystal structure of the calcium-dependent kinase from toxoplasma gondii, 541.m00134, kinase domain. | | Descriptor: | Calmodulin-like domain protein kinase isoform 3 | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Cossar, D, Wasney, G, Lin, Y.H, Hassani, A, Ali, A, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wikstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-24 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of the calcium-dependent kinase from toxoplasma gondii, 541.m00134, kinase domain.
To be Published
|
|
3DQN
 
 | |
2OLK
 
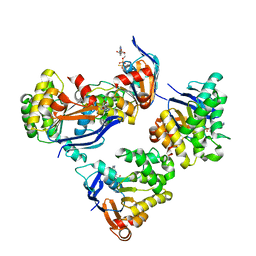 | | ABC Protein ArtP in complex with ADP-beta-S | | Descriptor: | 5'-O-[(R)-HYDROXY(THIOPHOSPHONOOXY)PHOSPHORYL]ADENOSINE, Amino acid ABC transporter | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the ATP-binding cassette (ABC) protein ArtP from Geobacillus stearothermophilus reveal a stable dimer in the post hydrolysis state and an asymmetry in the dimerization region
To be Published
|
|
4GH8
 
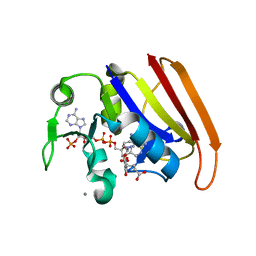 | | Crystal structure of a 'humanized' E. coli dihydrofolate reductase | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, METHOTREXATE, ... | | Authors: | French, J.B, Liu, C.T, Hanoian, P, Pringle, T.H, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2012-08-07 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional significance of evolving protein sequence in dihydrofolate reductase from bacteria to humans.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2OMT
 
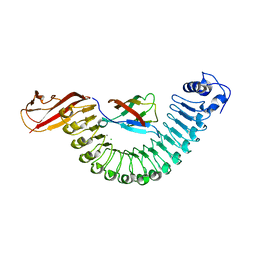 | | Crystal structure of InlA G194S+S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin; E-Cad/CTF1, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4ETU
 
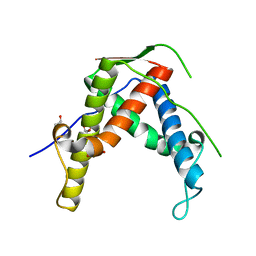 | |
3S9N
 
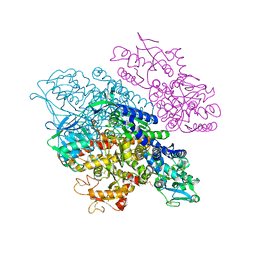 | | Complex between transferrin receptor 1 and transferrin with iron in the N-Lobe, room temperature | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eckenroth, B.E, Steere, A.N, Mason, A.B, Everse, S.J. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4EU7
 
 | |
4EUM
 
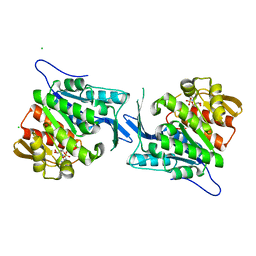 | | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II
To be Published
|
|
