5FVY
 
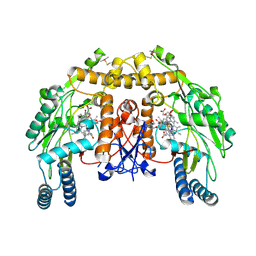 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 4-methyl-6-(2-(5-(4-methylpiperazin-1-yl)pyridin-3-yl) ethyl)pyridin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-6-(2-(5-(4-methylpiperazin-1-yl)pyridin-3-yl)ethyl)pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2016-02-10 | | Release date: | 2016-04-20 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibition by Optimization of the 2-Aminopyridine-Based Scaffold with a Pyridine Linker.
J.Med.Chem., 59, 2016
|
|
5FPA
 
 | |
5FVR
 
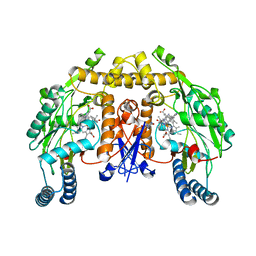 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-METHYL-6-(2-(5-(1-METHYLPIPERIDIN-4-YL)PYRIDIN-3-YL) ETHYL)PYRIDIN-2-AMINE | | Descriptor: | 4-methyl-6-(2-(5-(1-methylpiperidin-4-yl)pyridin-3-yl)ethyl)pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2016-02-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibition by Optimization of the 2-Aminopyridine-Based Scaffold with a Pyridine Linker.
J.Med.Chem., 59, 2016
|
|
1ETL
 
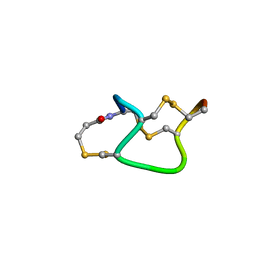 | |
1ETM
 
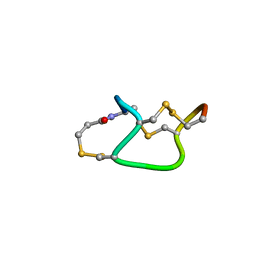 | |
7JJP
 
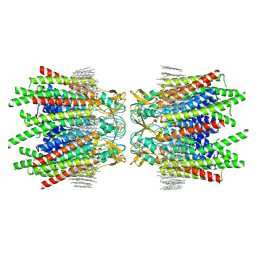 | | Sheep Connexin-50 at 1.9 angstroms resolution by CryoEM | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
1DZE
 
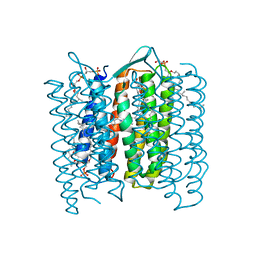 | | Structure of the M Intermediate of Bacteriorhodopsin trapped at 100K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Iizuka, T, Kamiya, N, Adachi, S, Kouyama, T. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the M Intermediate of Bacteriorhodopsin: Allosteric Structural Changes Mediated by Sliding Movement of a Transmembrane Helix
J.Mol.Biol., 341, 2004
|
|
1DXK
 
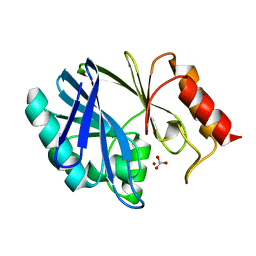 | |
5L8A
 
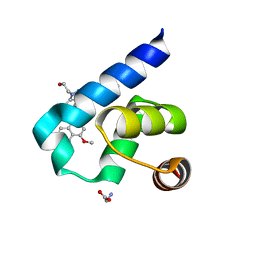 | | Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-hydroxyethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-~{N}-(phenylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, GLYCINE, ... | | Authors: | Dawidowski, M, Emmanouilidis, L, Sattler, M, Popowicz, G.M. | | Deposit date: | 2016-06-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.
Science, 355, 2017
|
|
7GRR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-14 | | Descriptor: | 3C-like proteinase nsp5, 5-(3-cyclohexylprop-1-yn-1-yl)pyridine-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6X7Q
 
 | | Chloramphenicol acetyltransferase type III in complex with chloramphenicol and acetyl-oxa(dethia)-CoA | | Descriptor: | CHLORAMPHENICOL, Chloramphenicol acetyltransferase 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-keto-acylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(de-thia)CoA
Acta Crystallogr.,Sect.F, 2023
|
|
1CFE
 
 | | P14A, NMR, 20 STRUCTURES | | Descriptor: | PATHOGENESIS-RELATED PROTEIN P14A | | Authors: | Fernandez, C, Szyperski, T, Bruyere, T, Ramage, P, Mosinger, E, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the pathogenesis-related protein P14a.
J.Mol.Biol., 266, 1997
|
|
5FLL
 
 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW) from Bacillus subtilis in complex with a Pimeloyl-adenylate | | Descriptor: | 6-CARBOXYHEXANOATE-COA LIGASE, MAGNESIUM ION, PIMELOYL-AMP, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-16 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
5I6E
 
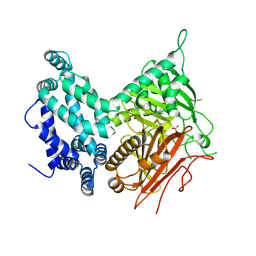 | | Crystal structure of the central domain of yeast acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase, MALONATE ION | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
5JH5
 
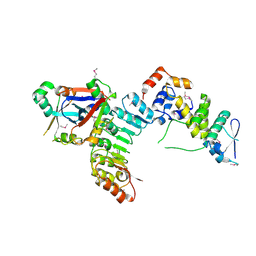 | | Structural Basis for the Hierarchical Assembly of the Core of PRC1.1 | | Descriptor: | BCL-6 corepressor-like protein 1, Lysine-specific demethylase 2B, Polycomb group RING finger protein 1, ... | | Authors: | Wong, S.J, Taylor, A.B, Hart, P.J, Kim, C.A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-09-14 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1.
Structure, 24, 2016
|
|
1ESD
 
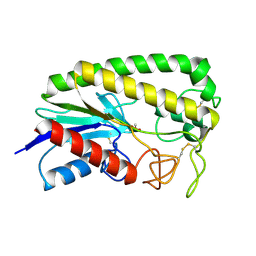 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | ESTERASE, METHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
1ESE
 
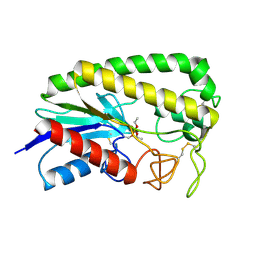 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | DIETHYL PHOSPHONATE, ESTERASE | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
1CKE
 
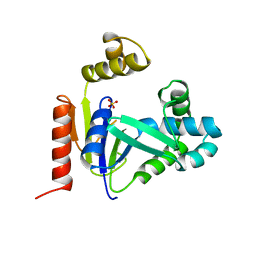 | | CMP KINASE FROM ESCHERICHIA COLI FREE ENZYME STRUCTURE | | Descriptor: | PROTEIN (CYTIDINE MONOPHOSPHATE KINASE), SULFATE ION | | Authors: | Briozzo, P, Golinelli-Pimpaneau, B. | | Deposit date: | 1998-09-24 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.
Structure, 6, 1998
|
|
5FDZ
 
 | | Crystal structure of human PCAF bromodomain in complex with compound BDOMB00091a (compound 14) | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase KAT2B, ~{N}-methyl-2-(oxan-4-yloxy)-5-(2-oxidanylidene-2-phenylazanyl-ethoxy)benzamide | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
1CQE
 
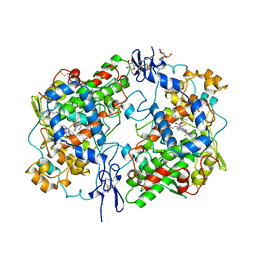 | | PROSTAGLANDIN H2 SYNTHASE-1 COMPLEX WITH FLURBIPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, PROTEIN (PROSTAGLANDIN H2 SYNTHASE-1), ... | | Authors: | Picot, D, Loll, P.J, Mulichak, A.M, Garavito, R.M. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1.
Nature, 367, 1994
|
|
1CTN
 
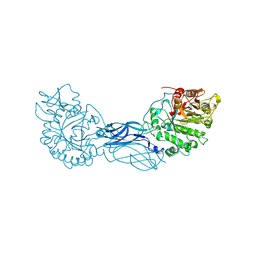 | | CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CHITINASE A | | Authors: | Perrakis, A, Tews, I, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial chitinase at 2.3 A resolution.
Structure, 2, 1994
|
|
7JM9
 
 | | Sheep Connexin-50 at 2.5 angstroms reoslution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7GLZ
 
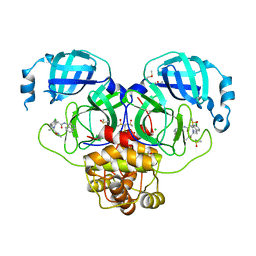 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-5cd9ea36-14 (Mpro-P2067) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(2S)-1-(methylamino)-1-oxopropan-2-yl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5HKK
 
 | | Caldalaklibacillus thermarum F1-ATPase (wild type) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Ferguson, S.A, Cook, G.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IFW
 
 | |
