1KOC
 
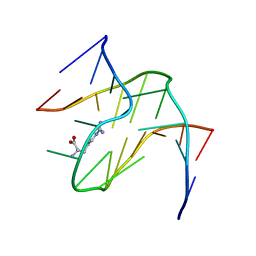 | | RNA APTAMER COMPLEXED WITH ARGININE, NMR | | Descriptor: | ARGININE, RNA (5'-R(P*AP*CP*AP*GP*GP*UP*AP*GP*GP*UP*CP*GP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*AP*GP*GP*AP*GP*CP*GP*U)-3') | | Authors: | Yang, Y.S, Kochoyan, M, Burgstaller, P, Westhof, E, Famulok, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy.
Science, 272, 1996
|
|
1LYX
 
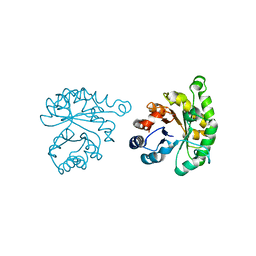 | | Plasmodium Falciparum Triosephosphate Isomerase (PfTIM)-Phosphoglycolate complex | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate Isomerase | | Authors: | Parthasarathy, S, Balaram, H, Balaram, P, Murthy, M.R. | | Deposit date: | 2002-06-10 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Plasmodium falciparum triosephosphate isomerase-phosphoglycolate complex in two crystal forms: characterization of catalytic loop open and closed conformations in the ligand-bound state
Biochemistry, 41, 2002
|
|
7XX0
 
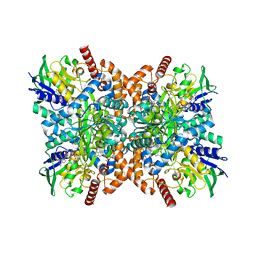 | | C281S glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XYL
 
 | | E426Q-glycine-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-06-01 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXM
 
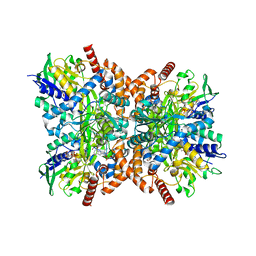 | | Orf1-glycine-4-aminobutylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
6WPE
 
 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 4-chloro-N-{[1-(3-chlorobenzene-1-carbonyl)-1,2,3,4-tetrahydroquinolin-6-yl]methyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Carbamate and N -Pyrimidine Mitigate Amide Hydrolysis: Structure-Based Drug Design of Tetrahydroquinoline IDO1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7XXD
 
 | | Orf1-sarcosine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-formimidoyl fortimicin A synthase, SARCOSINE | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7Y0X
 
 | | Orf1-glycine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-06-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XQA
 
 | | Orf1-glycine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXR
 
 | | Orf1 R342A-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-formimidoyl fortimicin A synthase, [(2~{R},3~{R},4~{S},5~{R},6~{R})-6-[(~{E})-[(3~{a}~{S},7~{R},7~{a}~{S})-7-oxidanyl-4-oxidanylidene-3,3~{a},5,6,7,7~{a}-hexahydro-1~{H}-imidazo[4,5-c]pyridin-2-ylidene]amino]-5-(2-azanylethanoylamino)-2-(hydroxymethyl)-4-oxidanyl-oxan-3-yl] carbamate | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXP
 
 | | F316A-glycine-streptothricin F complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
2AHX
 
 | | Crystal structure of ErbB4/HER4 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-4, SULFATE ION, ... | | Authors: | Bouyain, S, Longo, P.A, Li, S, Ferguson, K.M, Leahy, D.J. | | Deposit date: | 2005-07-28 | | Release date: | 2005-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4MLP
 
 | |
7CUM
 
 | | Cryo-EM structure of human GABA(B) receptor bound to the antagonist CGP54626 | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, CHOLESTEROL, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
5CUH
 
 | | Crystal structure MMP-9 complexes with a constrained hydroxamate based inhibitor LT4 | | Descriptor: | (4S)-3-{[4-(4-cyano-2-methylphenyl)piperazin-1-yl]sulfonyl}-N-hydroxy-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Tepshi, L, Vera, L, Nuti, E, Rosalia, L, Rossello, A, Stura, E.A. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a new selective inhibitor of A Disintegrin And Metalloprotease 10 (ADAM-10) able to reduce the shedding of NKG2D ligands in Hodgkin's lymphoma cell models.
Eur.J.Med.Chem., 111, 2016
|
|
8BPJ
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(D-p-FPhF)(O2CCH3)3] (Structure 1) | | Descriptor: | 9,11-bis(4-fluorophenyl)-3,7-dimethyl-2,4,6,8-tetraoxa-9,11-diaza-1$l^{4},5$l^{4}-diruthenatricyclo[3.3.3.0^{1,5}]undecane, Lysozyme, NITRATE ION, ... | | Authors: | Teran, A, Merlino, A, Ferraro, G. | | Deposit date: | 2023-01-17 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Effect of Equatorial Ligand Substitution on the Reactivity with Proteins of Paddlewheel Diruthenium Complexes: Structural Studies.
Inorg.Chem., 62, 2023
|
|
8BPU
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(D-p-FPhF)(O2CCH3)3] (Structure 2) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 9,11-bis(4-fluorophenyl)-2,4,6,8-tetraoxa-9,11-diaza-1$l^{4},5$l^{4}-diruthenatricyclo[3.3.3.0^{1,5}]undecane, Lysozyme, ... | | Authors: | Teran, A, Merlino, A, Ferraro, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Effect of Equatorial Ligand Substitution on the Reactivity with Proteins of Paddlewheel Diruthenium Complexes: Structural Studies.
Inorg.Chem., 62, 2023
|
|
8BPH
 
 | |
8BQM
 
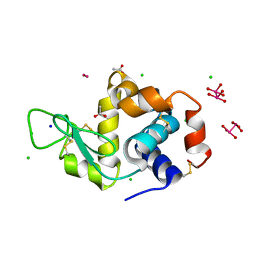 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(D-p-FPhF)(O2CCH3)3] (Structure 4) | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme, ... | | Authors: | Teran, A, Merlino, A, Ferraro, G. | | Deposit date: | 2022-11-21 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Effect of Equatorial Ligand Substitution on the Reactivity with Proteins of Paddlewheel Diruthenium Complexes: Structural Studies.
Inorg.Chem., 62, 2023
|
|
9D11
 
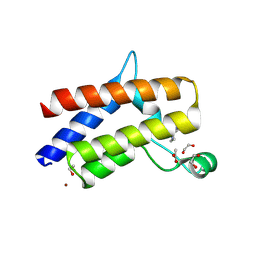 | | Smarca2 Bromodomain in complex with compound 22 | | Descriptor: | (12'S)-4'-chloro-10'-(piperidin-4-yl)-5'H-spiro[cyclohexane-1,7'-indolo[1,2-a]quinazolin]-5'-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2024-08-07 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Discovery of High-Affinity SMARCA2/4 Bromodomain Ligands and Development of Potent and Exceptionally Selective SMARCA2 PROTAC Degraders.
J.Med.Chem., 68, 2025
|
|
9D12
 
 | | Smarca2 Bromodomain in complex with compound 15 | | Descriptor: | (12'R)-4'-chloro-9'-(piperidin-4-yl)-5'H-spiro[cyclohexane-1,7'-indolo[1,2-a]quinazolin]-5'-one, ACETATE ION, Isoform Short of Probable global transcription activator SNF2L2, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2024-08-07 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Discovery of High-Affinity SMARCA2/4 Bromodomain Ligands and Development of Potent and Exceptionally Selective SMARCA2 PROTAC Degraders.
J.Med.Chem., 68, 2025
|
|
6G0D
 
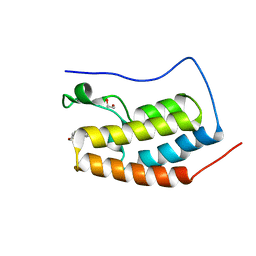 | |
7W8K
 
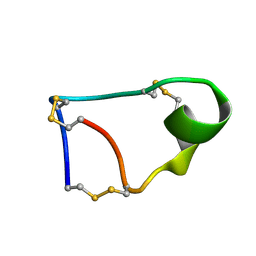 | |
7W8R
 
 | |
