258D
 
 | | FACTORS AFFECTING SEQUENCE SELECTIVITY ON NOGALAMYCIN INTERCALATION: THE CRYSTAL STRUCTURE OF D(TGTACA)-NOGALAMYCIN | | Descriptor: | ACETATE ION, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3'), NOGALAMYCIN, ... | | Authors: | Smith, C.K, Brannigan, J.A, Moore, M.H. | | Deposit date: | 1996-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Factors affecting DNA sequence selectivity of nogalamycin intercalation: the crystal structure of d(TGTACA)2-nogalamycin2.
J.Mol.Biol., 263, 1996
|
|
6S1C
 
 | | P3221 crystal form of the Ctf18-1-8/Pol2(1-528) complex | | Descriptor: | Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Grabarczyk, D.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (6.1 Å) | | Cite: | Ctf18-RFC and DNA Pol ε form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication.
Nucleic Acids Res., 48, 2020
|
|
1Q3C
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli: The E2A mutant at 2.3 resolution. | | Descriptor: | Endonuclease VIII, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Golan, G, Zharkov, D.O, Feinberg, H, Fernandes, A.S, Zaika, E.I, Kycia, J.H, Grollman, A.P, Shoham, G. | | Deposit date: | 2003-07-29 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.
Nucleic Acids Res., 33, 2005
|
|
2Y7H
 
 | | Atomic model of the DNA-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | 5'-D(*GP*TP*TP*CP*AP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*GP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*CP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*TP*GP*AP*AP*C)-3', S-ADENOSYLMETHIONINE, ... | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
1WTB
 
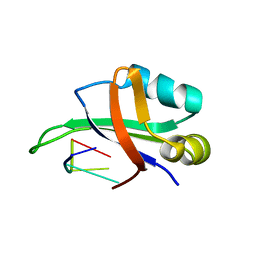 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D (AUF1) with telomere DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2004-11-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D
J.Biol.Chem., 280, 2005
|
|
1ZR2
 
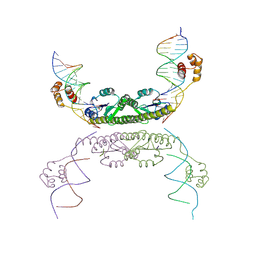 | | Structure of a Synaptic gamma-delta Resolvase Tetramer Covalently Linked to two Cleaved DNAs | | Descriptor: | AAA, TCAGTGTCCGATAATTTAT, TTATCGGACACTG, ... | | Authors: | Li, W, Kamtekar, S, Xiong, Y, Sarkis, G.J, Grindley, N.D, Steitz, T.A. | | Deposit date: | 2005-05-18 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs.
Science, 309, 2005
|
|
1X0F
 
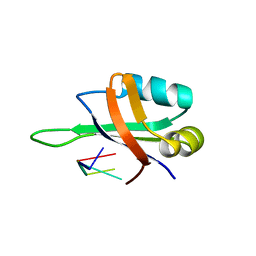 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D(AUF1) with telomeric DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2005-03-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D.
J.Biol.Chem., 280, 2005
|
|
3S5A
 
 | | ABH2 cross-linked to undamaged dsDNA-2 with cofactors | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*CP*AP*CP*TP*GP*TP*CP*G)-3', 5'-D(*TP*CP*GP*AP*CP*AP*GP*TP*GP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Ramirez, B, Zhang, W, Jia, G, Zhang, L, Li, C.Q, Dinner, A.R, Yang, C.-G, He, C. | | Deposit date: | 2011-05-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3QN2
 
 | |
8FAY
 
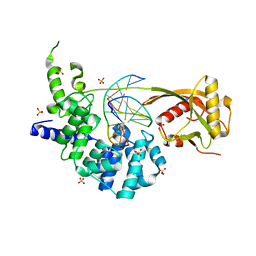 | | Human MUTYH adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*CP*AP*(NR1)P*GP*TP*CP*T)-3'), ... | | Authors: | Trasvina-Arenas, C.H, Lin, W.J, Demir, M, Fisher, A.J, David, S.S, Horvath, M.P. | | Deposit date: | 2022-11-29 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Human MUTYH adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G)
To Be Published
|
|
1M6R
 
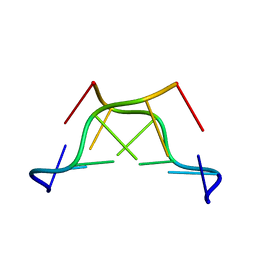 | |
1ZR4
 
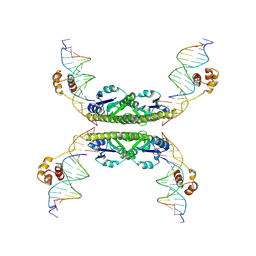 | | Structure of a Synaptic gamma-delta Resolvase Tetramer Covalently linked to two Cleaved DNAs | | Descriptor: | AAA, TCAGTGTCCGATAATTTAT, TTATCGGACACTG, ... | | Authors: | Li, W, Kamtekar, S, Xiong, Y, Sarkis, G.J, Grindley, N.D, Steitz, T.A. | | Deposit date: | 2005-05-19 | | Release date: | 2005-08-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs.
Science, 309, 2005
|
|
1OYI
 
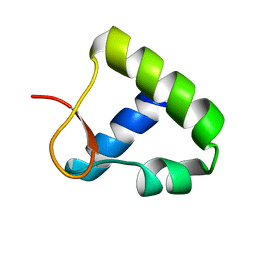 | | Solution structure of the Z-DNA binding domain of the vaccinia virus gene E3L | | Descriptor: | double-stranded RNA-binding protein | | Authors: | Kahmann, J.D, Wecking, D.A, Putter, V, Lowenhaupt, K, Kim, Y.-G, Schmieder, P, Oschkinat, H, Rich, A, Schade, M. | | Deposit date: | 2003-04-04 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8UK4
 
 | | Crystal structure of human polymerase eta with incoming dTMPnPP nucleotide opposite urea lesion | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(UD8)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Tomar, R, Egli, M, Stone, M.P. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Replication Bypass of the N -(2-Deoxy-d-erythro-pentofuranosyl)-urea DNA Lesion by Human DNA Polymerase eta.
Biochemistry, 63, 2024
|
|
8UJV
 
 | | Crystal structure of human polymerase eta with incoming dCMPnPP nucleotide opposite urea lesion | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(UD8)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Tomar, R, Egli, M, Stone, M.P. | | Deposit date: | 2023-10-11 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Replication Bypass of the N -(2-Deoxy-d-erythro-pentofuranosyl)-urea DNA Lesion by Human DNA Polymerase eta.
Biochemistry, 63, 2024
|
|
8UJX
 
 | | Crystal structure of human polymerase eta with incoming dGMPnPP nucleotide opposite urea lesion | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(UD8)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Tomar, R, Egli, M, Stone, M.P. | | Deposit date: | 2023-10-11 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Replication Bypass of the N -(2-Deoxy-d-erythro-pentofuranosyl)-urea DNA Lesion by Human DNA Polymerase eta.
Biochemistry, 63, 2024
|
|
7V0C
 
 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, MANGANESE (II) ION, Palindromic DNA18, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG
To Be Published
|
|
7V0R
 
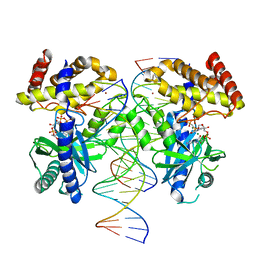 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -ppcpG(2 ,5 )pA | | Descriptor: | Cyclic GMP-AMP synthase, MAGNESIUM ION, Palindromic DNA18, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -ppcpG(2 ,5 )pA
To Be Published
|
|
7N3Z
 
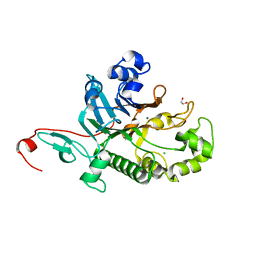 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-(apurinic or apyrimidinic site) endonuclease 2, ... | | Authors: | Wojtaszek, J.L, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
2VHG
 
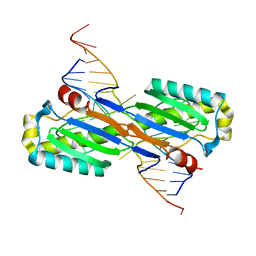 | | Crystal Structure of the ISHp608 Transposase in Complex with Right End 31-mer DNA | | Descriptor: | MANGANESE (II) ION, RIGHT END 31-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-11-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
5EUG
 
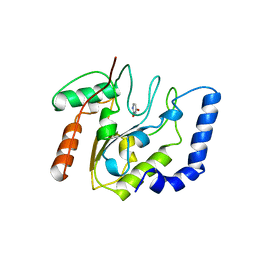 | | CRYSTALLOGRAPHIC AND ENZYMATIC STUDIES OF AN ACTIVE SITE VARIANT H187Q OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE: CRYSTAL STRUCTURES OF MUTANT H187Q AND ITS URACIL COMPLEX | | Descriptor: | PROTEIN (GLYCOSYLASE), URACIL | | Authors: | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-12-27 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
4L5E
 
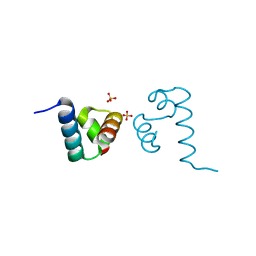 | | Crystal structure of A. aeolicus NtrC1 DNA binding domain | | Descriptor: | SULFATE ION, Transcriptional regulator (NtrC family) | | Authors: | Young, A, Maris, A.E, Vidangos, N.K, Hong, E, Pelton, J.G, Batchelor, J.D, Wemmer, D.E. | | Deposit date: | 2013-06-10 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure, function, and tethering of DNA-binding domains in sigma (54) transcriptional activators.
Biopolymers, 99, 2013
|
|
3E41
 
 | | Q138F HincII bound to GTCGAC and 5 mM Ca2+ | | Descriptor: | 5'-D(*DGP*DCP*DCP*DGP*DGP*DTP*DCP*DGP*DAP*DCP*DCP*DGP*DGP*DC)-3', CALCIUM ION, SODIUM ION, ... | | Authors: | Horton, N.C, Babic, A.C, Little, E.J, Manohar, V.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | DNA distortion and specificity in a sequence-specific endonuclease.
J.Mol.Biol., 383, 2008
|
|
5EW1
 
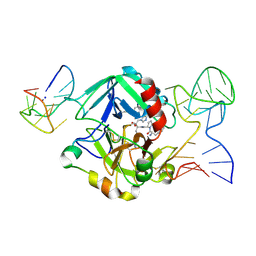 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT3, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
2VJU
 
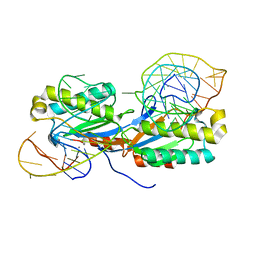 | | Crystal structure of the IS608 transposase in complex with the complete Right end 35-mer DNA and manganese | | Descriptor: | MANGANESE (II) ION, RIGHT END 35-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
