6V4S
 
 | |
6T60
 
 | |
6T6N
 
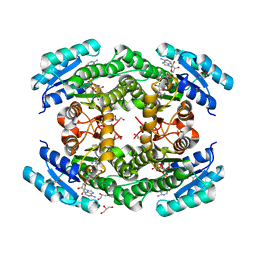 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) in complex with NADH at 2.5 A resolution | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-[acyl-carrier protein] reductase, D-MALATE, ... | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
3Q2W
 
 | | Crystal structure of mouse N-cadherin ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
6VDE
 
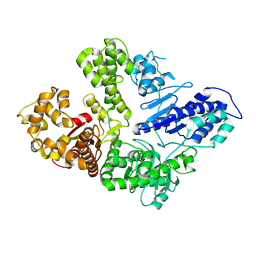 | | Full-length M. smegmatis Pol1 | | Descriptor: | DNA polymerase I, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
3JSS
 
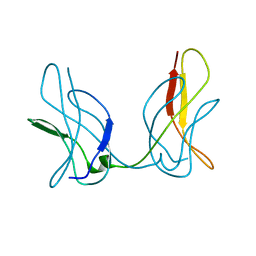 | |
6T77
 
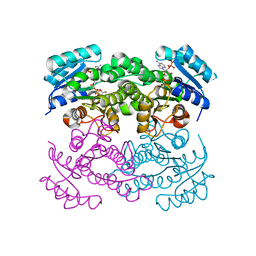 | | Crystal structure of Klebsiella pneumoniae FabG(NADPH-dependent) NADP-complex at 1.75 A resolution | | Descriptor: | 3-oxoacyl-ACP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
3QF2
 
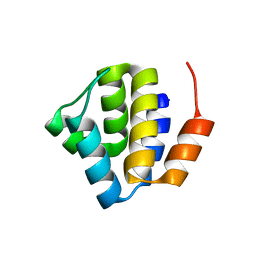 | | Crystal structure of NALP3 PYD | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Park, H.H, Bae, J.Y. | | Deposit date: | 2011-01-21 | | Release date: | 2011-08-31 | | Last modified: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of NALP3 Protein Pyrin Domain (PYD) and Its Implications in Inflammasome Assembly
J.Biol.Chem., 286, 2011
|
|
5EMS
 
 | | Crystal Structure of an iodinated insulin analog | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Lawrence, M.C, Pandyarajan, V, Wan, Z, Weiss, M.A. | | Deposit date: | 2015-11-06 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extending Halogen-based Medicinal Chemistry to Proteins: IODO-INSULIN AS A CASE STUDY.
J. Biol. Chem., 291, 2016
|
|
8XWX
 
 | |
6K6I
 
 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens | | Descriptor: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens
To Be Published
|
|
2AJN
 
 | | NMR structure of the in-plane membrane anchor domain [1-28] of the monotopic NonStructural Protein 5A (NS5A) from the Bovine Viral Diarrhea Virus (BVDV) | | Descriptor: | Nonstructural protein 5A | | Authors: | Sapay, N, Montserret, R, Chipot, C, Brass, V, Moradpour, D, Deleage, G, Penin, F. | | Deposit date: | 2005-08-02 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus.
Biochemistry, 45, 2006
|
|
6K6K
 
 | | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens | | Descriptor: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens
To Be Published
|
|
1CH4
 
 | | MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA (F133V) | | Descriptor: | CARBON MONOXIDE, MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shirai, T, Fujikake, M, Yamane, T, Inaba, K, Ishimori, K, Morishima, I. | | Deposit date: | 1998-06-11 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a protein with an artificial exon-shuffling, module M4-substituted chimera hemoglobin beta alpha, at 2.5 A resolution.
J.Mol.Biol., 287, 1999
|
|
6K6J
 
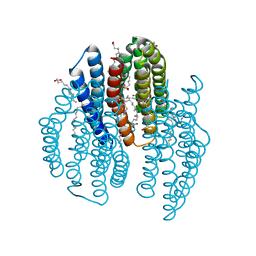 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion | | Descriptor: | BROMIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion
To Be Published
|
|
1SQE
 
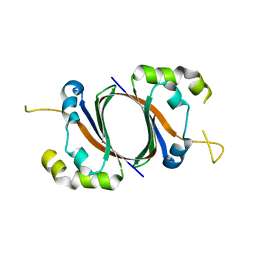 | | 1.5A Crystal Structure Of the protein PG130 from Staphylococcus aureus, Structural genomics | | Descriptor: | hypothetical protein PG130 | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases
J.Biol.Chem., 280, 2005
|
|
3Q2N
 
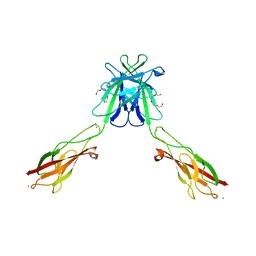 | | Mouse E-cadherin EC1-2 L175D mutant | | Descriptor: | CALCIUM ION, Cadherin-1, TETRAETHYLENE GLYCOL | | Authors: | Harrison, O.J, Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
1T3G
 
 | |
2DYU
 
 | |
2E2K
 
 | |
3QF4
 
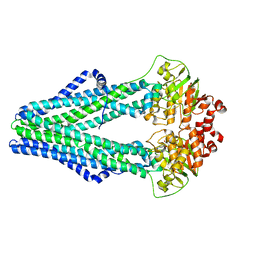 | | Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation | | Descriptor: | ABC transporter, ATP-binding protein, MAGNESIUM ION, ... | | Authors: | Hohl, M, Briand, C, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation
Nat.Struct.Mol.Biol., 19, 2012
|
|
3Q2L
 
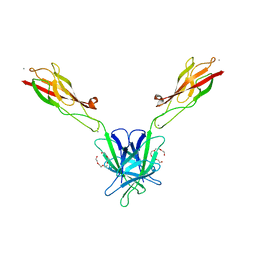 | | Mouse E-cadherin EC1-2 V81D mutant | | Descriptor: | CALCIUM ION, Cadherin-1, PENTAETHYLENE GLYCOL | | Authors: | Harrison, O.J, Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
2NA0
 
 | |
2DYV
 
 | |
8X8Q
 
 | | Structure of enterovirus protease in complex host factor | | Descriptor: | 2A protein (Fragment), Actin-histidine N-methyltransferase, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2023-11-28 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
