7AS3
 
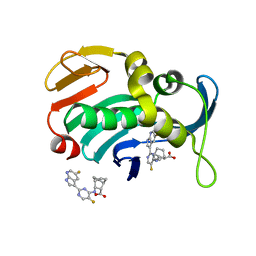 | | Influenza A PB2 (H357N mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7B2U
 
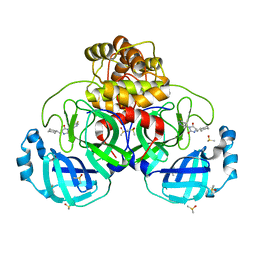 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 1 | | Descriptor: | (5S)-5-(cyclohexylmethyl)-3-(5-fluoropyridin-3-yl)imidazolidine-2,4-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B5Z
 
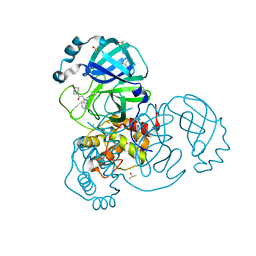 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 6 | | Descriptor: | 2-(1H-benzo[d][1,2,3]triazol-1-yl)-1-(4-methylenepiperidin-1-yl)ethan-1-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-12-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
3CL9
 
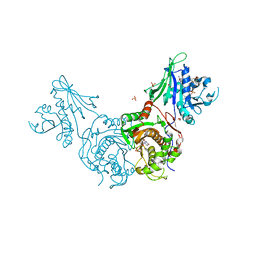 | | Structure of bifunctional TcDHFR-TS in complex with MTX | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS), ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
5M3Q
 
 | |
5MVC
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 4-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,5-thiadiazole-3-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
7B2J
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 5 | | Descriptor: | 2-(1H-1,2,3-benzotriazol-1-yl)-1-(4-methylpiperidin-1-yl)ethan-1-one, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7AU4
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 3 | | Descriptor: | (3~{S})-6-chloranyl-3'-(1,2-oxazol-3-ylmethyl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-02 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B77
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 8 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-ethyl-~{N}-(furan-3-ylmethyl)ethanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-12-09 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
3CLB
 
 | | Structure of bifunctional TcDHFR-TS in complex with TMQ | | Descriptor: | 1,2-ETHANEDIOL, DHFR-TS, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
3UVE
 
 | |
3ZH3
 
 | |
5M43
 
 | |
5MUT
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 2-methyl-5-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,3-triazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
3FL5
 
 | | Protein kinase CK2 in complex with the inhibitor Quinalizarin | | Descriptor: | 1,2,5,8-tetrahydroxyanthracene-9,10-dione, Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER | | Authors: | Mazzorana, M, Franchin, C, Battistutta, R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quinalizarin as a potent, selective and cell-permeable inhibitor of protein kinase CK2
Biochem.J., 421, 2009
|
|
3FV7
 
 | | OXA-24 beta-lactamase complex with SA4-44 inhibitor | | Descriptor: | (2S)-2-[[2-methanoyl-7-(methoxycarbonylamino)indolizin-3-yl]amino]-3-methyl-3-sulfino-butanoic acid, Beta-lactamase OXA-24 | | Authors: | Bou, G, Santillana, E, Sheri, A, Beceiro, A, Sampson, J.M, Kalp, M, Bethel, C.R, Distler, A.M, Drawz, S.M, Pagadala, S.R, Van den Akker, F, Bonomo, R.A, Romero, A, Buynak, J.D. | | Deposit date: | 2009-01-15 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase.
J.Am.Chem.Soc., 132, 2010
|
|
3FYD
 
 | |
3FZC
 
 | | OXA-24 beta-lactamase complex with SA3-53 inhibitor | | Descriptor: | (2S,3R)-4-(2-aminoethylcarbamoyloxy)-2-[(2-methanoylindolizin-3-yl)amino]-3-methyl-3-sulfino-butanoic acid, Beta-lactamase OXA-24, TETRAETHYLENE GLYCOL | | Authors: | Romero, A, Santillana, E. | | Deposit date: | 2009-01-24 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase.
J.Am.Chem.Soc., 132, 2010
|
|
3FYZ
 
 | | OXA-24 beta-lactamase complex with SA4-17 inhibitor | | Descriptor: | (2S,3R)-2-[(7-aminocarbonyl-2-methanoyl-indolizin-3-yl)amino]-4-aminocarbonyloxy-3-methyl-3-sulfino-butanoic acid, Beta-lactamase OXA-24, TETRAETHYLENE GLYCOL | | Authors: | Romero, A, Santillana, E. | | Deposit date: | 2009-01-23 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase.
J.Am.Chem.Soc., 132, 2010
|
|
3ZS6
 
 | | The Structural characterization of Burkholderia pseudomallei OppA. | | Descriptor: | CHLORIDE ION, GLYCEROL, OLIGOPEPTIDE DVA, ... | | Authors: | Lassaux, P, Gourlay, L.J, Bolognesi, M. | | Deposit date: | 2011-06-23 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Structure-Based Strategy for Epitope Discovery in Burkholderia Pseudomallei Oppa Antigen.
Structure, 21, 2013
|
|
3ZH4
 
 | |
3G4P
 
 | | OXA-24 beta-lactamase at pH 7.5 | | Descriptor: | Beta-lactamase OXA-24 | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2009-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase
J.Am.Chem.Soc., 132, 2010
|
|
3FYC
 
 | |
3DC1
 
 | | Crystal structure of kynurenine aminotransferase II complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Kynurenine/alpha-aminoadipate aminotransferase mitochondrial | | Authors: | Han, Q, Cai, T, Tagle, D.A, Robinson, H, Li, J. | | Deposit date: | 2008-06-03 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
Biosci.Rep., 28, 2008
|
|
4B5C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia pseudomallei | | Descriptor: | ACETATE ION, PUTATIVE OMPA FAMILY LIPOPROTEIN | | Authors: | Gourlay, L.J, Peri, C, Conchillo-Sole, O, Ferrer-Navarro, M, Gori, A, Longhi, R, Rinchai, D, Lertmemongkolchai, G, Lassaux, P, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Burkholderia Pseudomallei Acute Phase Antigen Bpsl2765 for Structure-Based Epitope Discovery/Design in Structural Vaccinology.
Chem.Biool., 20, 2013
|
|
