4W9O
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2PPO
 
 | |
8CCG
 
 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12S)-12-(thiophen-2-yl)-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15-dimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{S})-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15-dimethyl-12-thiophen-2-yl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
5HUA
 
 | | Structure of C. glabrata FKBP12-FK506 complex | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-01-27 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of Pathogenic Fungal FKBP12s Reveal Possible Self-Catalysis Function.
Mbio, 7, 2016
|
|
3O5L
 
 | | Fk1 domain mutant A19T of FKBP51, crystal form I | | Descriptor: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5DIU
 
 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand 2-(3-((R)-1-((S)-1-((S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)acetyl)piperidine-2-carboxamido)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-1-[({(2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)acetyl]piperidin-2-yl}carbonyl)amino]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetic acid | | Authors: | Gaali, S, Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Rapid, Structure-Based Exploration of Pipecolic Acid Amides as Novel Selective Antagonists of the FK506-Binding Protein 51.
J.Med.Chem., 59, 2016
|
|
3UQI
 
 | |
7ETV
 
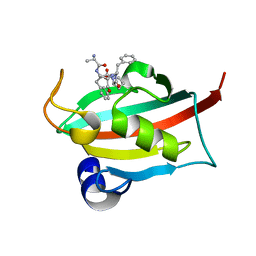 | | The FK1 domain of FKBP51 in complex with peptide-inhibitor hit DFPFV | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, peptide-inhibitor hit | | Authors: | Han, J.T, Zhu, Y.C, Pan, D.B, Xue, H.X, Wang, S, Liu, H.X, He, Y.X, Yao, X.J. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Discovery of pentapeptide-inhibitor hits targeting FKBP51 by combining computational modeling and X-ray crystallography.
Comput Struct Biotechnol J, 19, 2021
|
|
8CCA
 
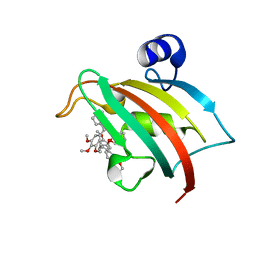 | | The Fk1 domain of FKBP51 in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
8BK4
 
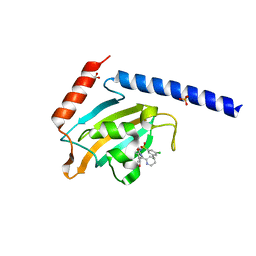 | | Full length structure of the apo-state LpMIP. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-[(1~{S})-1-pyridin-2-ylethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, GLYCEROL, Macrophage infectivity potentiator, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
7B9Y
 
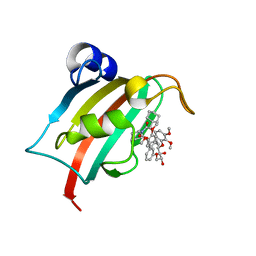 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 64a | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,24(28),25-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
4DT4
 
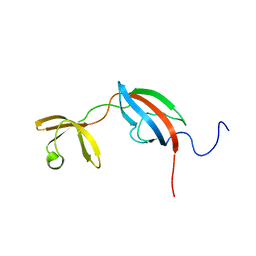 | |
7U8D
 
 | | FKBP12 mutant V55G bound to Rapa*-3Z | | Descriptor: | (3S,5Z,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-5-(ethoxyimino)-9,27-dihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Wassarman, D.R, Shokat, K.M. | | Deposit date: | 2022-03-08 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Tissue-restricted inhibition of mTOR using chemical genetics.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ETU
 
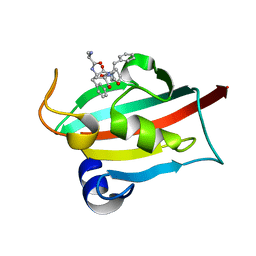 | | The FK1 domain of FKBP51 in complex with peptide-inhibitor hit SFPFT | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, peptide-inhibitor hit | | Authors: | Han, J.T, Zhu, Y.C, Pan, D.B, Xue, H.X, Wang, S, Liu, H.X, He, Y.X, Yao, X.J. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery of pentapeptide-inhibitor hits targeting FKBP51 by combining computational modeling and X-ray crystallography.
Comput Struct Biotechnol J, 19, 2021
|
|
8CCE
 
 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)butanoyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)butanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
4TW6
 
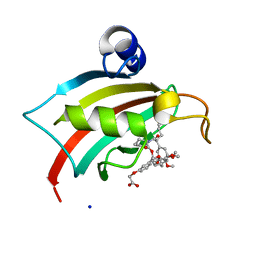 | | The Fk1 domain of FKBP51 in complex with iFit1 | | Descriptor: | (3-{(1R)-3-(3,4-dimethoxyphenyl)-1-[({(2S)-1-[(2S)-2-(3,4,5-trimethoxyphenyl)pent-4-enoyl]piperidin-2-yl}carbonyl)oxy]propyl}phenoxy)acetic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
8CHL
 
 | | Human FKBP12 in complex with (1S,5S,6R)-9-((3,5-dichlorophenyl)sulfonyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,9-diazabicyclo[4.2.1]nonan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, CADMIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
7AWF
 
 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{R})-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-11-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4QT3
 
 | | Crystal structure resolution of Plasmodium falciparum FK506 binding domain (FKBP35) in complex with Rapamycin at 1.4A resolution | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, FK506-binding protein (FKBP)-type peptidyl-propyl isomerase, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Bianchin, A, Allemand, F, Bell, A, Chubb, A.J, Guichou, J.-F. | | Deposit date: | 2014-07-07 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two crystal structures of the FK506-binding domain of Plasmodium falciparum FKBP35 in complex with rapamycin at high resolution
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ODK
 
 | |
6TX9
 
 | |
3NI6
 
 | |
7B9Z
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 35-(E) | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,20,24(28),25-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5, isothiocyanate | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8BK5
 
 | | A structure of the truncated LpMIP with bound inhibitor JK095. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase, SODIUM ION | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
4QT2
 
 | | Crystal Structure of the FK506-Binding Domain of Plasmodium Falciparum FKBP35 in complex with Rapamycin | | Descriptor: | FK506-binding protein (FKBP)-type peptidyl-propyl isomerase, IMIDAZOLE, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Bianchin, A, Allemand, F, Bell, A, Chubb, A.J, Guichou, J.-F. | | Deposit date: | 2014-07-07 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Two crystal structures of the FK506-binding domain of Plasmodium falciparum FKBP35 in complex with rapamycin at high resolution
Acta Crystallogr.,Sect.D, 71, 2015
|
|
