5KK3
 
 | | Atomic Resolution Structure of Monomorphic AB42 Amyloid Fibrils | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Colvin, M.T, Silvers, R, Zhe Ni, Q, Can, T.V, Sergeyev, I, Rosay, M, Donovan, K.J, Michael, B, Wall, J, Linse, S, Griffin, R.G. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic Resolution Structure of Monomorphic A beta 42 Amyloid Fibrils.
J.Am.Chem.Soc., 138, 2016
|
|
1TEX
 
 | | Mycobacterium smegmatis Stf0 Sulfotransferase with Trehalose | | Descriptor: | Stf0 Sulfotransferase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Mougous, J.D, Petzold, C.J, Senaratne, R.H, Lee, D.H, Akey, D.L, Lin, F.L, Munchel, S.E, Pratt, M.R, Riley, L.W, Leary, J.A, Berger, J.M, Bertozzi, C.R. | | Deposit date: | 2004-05-25 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1TFF
 
 | | Structure of Otubain-2 | | Descriptor: | Ubiquitin thiolesterase protein OTUB2 | | Authors: | Nanao, M.H, Tcherniuk, S.O, Chroboczek, J, Dideberg, O, Dessen, A, Balakirev, M.Y. | | Deposit date: | 2004-05-27 | | Release date: | 2004-08-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human otubain 2.
Embo Rep., 5, 2004
|
|
1TFQ
 
 | | NMR Structure of an Antagonists of the XIAP-Caspase-9 Interaction Complexed to the BIR3 domain of XIAP | | Descriptor: | Baculoviral IAP repeat-containing protein 4, N-METHYLALANYL-3-METHYLVALYL-N-(1,2,3,4-TETRAHYDRONAPHTHALEN-1-YL)PROLINAMIDE, ZINC ION | | Authors: | Oost, T.K, Sun, C, Armstrong, R.C, Al-Assaad, A.S, Betz, S.F, Deckwerth, T.L, Elmore, S.W, Meadows, R.P, Olejniczak, E.T, Oleksijew, A, Oltersdorf, T, Rosenberg, S.H, Shoemaker, A.R, Zou, H, Fesik, S.W. | | Deposit date: | 2004-05-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Discovery of Potent Antagonists of the Antiapoptotic Protein XIAP for the Treatment of Cancer.
J.Med.Chem., 47, 2004
|
|
4DD9
 
 | | EVAL processed HEWL, carboplatin DMSO paratone | | Descriptor: | DIMETHYL SULFOXIDE, Lysozyme C, carboplatin | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5KSX
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor AM-02-072 | | Descriptor: | Farnesyl pyrophosphate synthase, PHOSPHATE ION, [[(2~{S})-2-[[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoyl]amino]phosphonic acid | | Authors: | Park, J, Matralis, A, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-07-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
3L2P
 
 | | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching Between Two DNA Bound States | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*CP*CP*G)-3', 5'-D(*GP*TP*CP*GP*GP*AP*CP*TP*G)-3', 5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*C)-3', ... | | Authors: | Cotner-Gohara, E.A, Kim, I.K, Hammel, M, Tainer, J.A, Tomkinson, A, Ellenberger, T. | | Deposit date: | 2009-12-15 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching between Two DNA-Bound States.
Biochemistry, 49, 2010
|
|
3KN1
 
 | | Crystal Structure of Golgi Phosphoprotein 3 N-term Truncation Variant | | Descriptor: | Golgi phosphoprotein 3, SULFATE ION | | Authors: | Schmitz, K.R, Bessman, N.J, Setty, T.G, Ferguson, K.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking.
J.Cell Biol., 187, 2009
|
|
1TK0
 
 | | T7 DNA polymerase ternary complex with 8 oxo guanosine and ddCTP at the insertion site | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*CP*CP*CP*(8OG)P*CP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-07 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
5UKQ
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522.2 Fab | | Descriptor: | DH522.2 Fab fragment heavy chain, DH522.2 Fab fragment light chain, GLYCEROL | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
1T1L
 
 | | Crystal structure of the long-chain fatty acid transporter FadL | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Long-chain fatty acid transport protein | | Authors: | van den Berg, B, Black, P.N, Clemons Jr, W.M, Rapoport, T.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the long-chain fatty acid transporter FadL.
Science, 304, 2004
|
|
1UXE
 
 | | ADENOVIRUS AD37 FIBRE HEAD | | Descriptor: | ACETATE ION, FIBER PROTEIN, ZINC ION | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
3IE4
 
 | | b-glucan binding domain of Drosophila GNBP3 defines a novel family of pattern recognition receptor | | Descriptor: | 1,2-ETHANEDIOL, Gram-Negative Binding Protein 3, ZINC ION | | Authors: | Mishima, Y, Coste, F, Kellenberger, C, Roussel, A. | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The N-terminal domain of drosophila gram-negative binding protein 3 (GNBP3) defines a novel family of fungal pattern recognition receptors
To be Published
|
|
3IFN
 
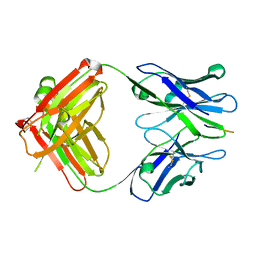 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-40:12A11) complex | | Descriptor: | 12A11 FAB antibody heavy chain, 12A11 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
1TBP
 
 | |
1TOK
 
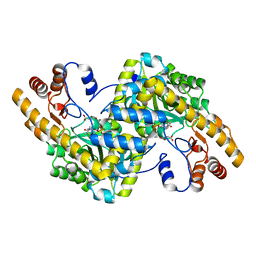 | | Maleic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, MALEIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOJ
 
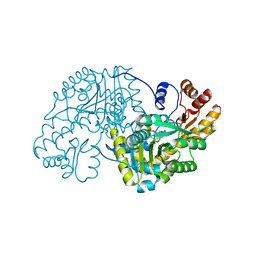 | | Hydrocinnamic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOO
 
 | | Interleukin 1B Mutant F146W | | Descriptor: | Interleukin-1 beta | | Authors: | Adamek, D.H, Guerrero, L, Caspar, D.L. | | Deposit date: | 2004-06-14 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and energetic consequences of mutations in a solvated hydrophobic cavity.
J.Mol.Biol., 346, 2005
|
|
1TP0
 
 | |
1TR7
 
 | | FimH adhesin receptor binding domain from uropathogenic E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, FimH protein, ... | | Authors: | Bouckaert, J, Berglund, J, Schembri, M, De Genst, E, Cools, L, Wuhrer, M, Hung, C.S, Pinkner, J, Slattegard, R, Zavialov, A, Choudhury, D, Langermann, S, Hultgren, S.J, Wyns, L, Klemm, P, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-06-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin
Mol.Microbiol., 55, 2005
|
|
1TWM
 
 | | Interleukin-1 Beta Mutant F146Y | | Descriptor: | Interleukin-1 beta | | Authors: | Adamek, D.H, Capsar, D.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and energetic consequences of mutations in a solvated hydrophobic cavity.
J.Mol.Biol., 346, 2005
|
|
1TXT
 
 | | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, ACETOACETYL-COENZYME A | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
1TOI
 
 | | Hydrocinnamic acid-bound structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
7YFN
 
 | | Core module of the NuA4 complex in S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3KN4
 
 | | AGAO 6-phenyl-2,3-hexadienylamine complex | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Nguyen, Y.H, Ernberg, K.E, Guss, J.M. | | Deposit date: | 2009-11-12 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | AGAO 6-phenyl-2,3-hexadienylamine complex
To be Published
|
|
