8YB7
 
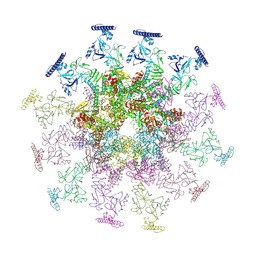 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C3 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8ZMR
 
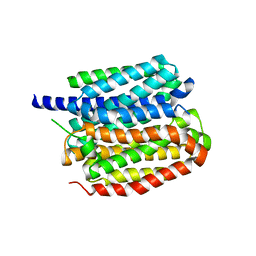 | | Vesamicol-bound VAChT | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7, vesamicol | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 34, 2024
|
|
8ZMS
 
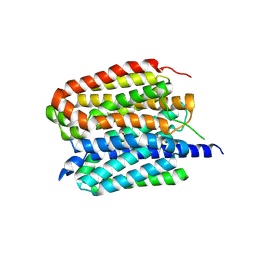 | | Acetylcholine-bound VAChT | | Descriptor: | ACETYLCHOLINE, Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7 | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 34, 2024
|
|
8YB5
 
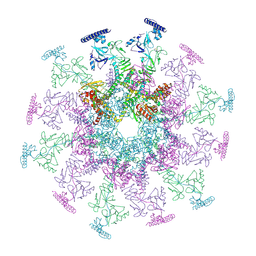 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C6 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8ZB8
 
 | | Crystal structure of T2R-TTL-DPP21 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Detyrosinated tubulin alpha-1B chain, ... | | Authors: | Wu, C.Y, Chen, J.J. | | Deposit date: | 2024-04-26 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of T2R-TTL-DPP21 complex
To Be Published
|
|
7P1N
 
 | | Crystal structure of human acetylcholinesterase in complex with (2R,3R,4S,5S,6R)-2-{4-[1-(4-{5-hydroxy-6-[(E)-(hydroxyimino)methyl]pyridin-2-yl}butyl)-1H-1,2,3-triazol-4-yl]butoxy}-6-(hydroxymethyl)oxane-3,4,5-triol oxime | | Descriptor: | (2R,3R,4S,5S,6R)-2-[4-[1-[4-[6-[(Z)-hydroxyiminomethyl]-5-oxidanyl-pyridin-2-yl]butyl]-1,2,3-triazol-4-yl]butoxy]-6-(hydroxymethyl)oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Da Silva, O, Dias, J, Nachon, F. | | Deposit date: | 2021-07-02 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A New Class of Bi- and Trifunctional Sugar Oximes as Antidotes against Organophosphorus Poisoning.
J.Med.Chem., 65, 2022
|
|
9B60
 
 | |
9B5Z
 
 | |
9B64
 
 | |
9B67
 
 | |
9B6A
 
 | |
9B63
 
 | |
9B61
 
 | |
7OL2
 
 | | Crystal structure of mouse contactin 1 immunoglobulin domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, ... | | Authors: | Chataigner, L.M.P, Janssen, B.J.C. | | Deposit date: | 2021-05-19 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structural insights into the contactin 1 - neurofascin 155 adhesion complex.
Nat Commun, 13, 2022
|
|
7OL4
 
 | |
7OK5
 
 | | Crystal structure of mouse neurofascin 155 immunoglobulin domains | | Descriptor: | Neurofascin 155, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chataigner, L.M.P, Janssen, B.J.C. | | Deposit date: | 2021-05-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into the contactin 1 - neurofascin 155 adhesion complex.
Nat Commun, 13, 2022
|
|
7PSG
 
 | | Structure of the ligand binding domain of the PacA (ECA2226) chemoreceptor of Pectobacterium atrosepticum SCRI1043 in complex with betaine. | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, TRIMETHYL GLYCINE | | Authors: | Gavira, J.A, Matilla, M.A, Velando, F, Krell, T. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
7PRR
 
 | | Structure of the ligand binding domain of the PctD (PA4633) chemoreceptor of Pseudomonas aeruginosa PAO1 in complex with acetylcholine | | Descriptor: | ACETYLCHOLINE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Matilla, M.A, Martin-Mora, D, Krell, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
7PRQ
 
 | | Structure of the ligand binding domain of the PctD (PA4633) chemoreceptor of Pseudomonas aeruginosa PAO1 in complex with choline. | | Descriptor: | 1,2-ETHANEDIOL, CHOLINE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Matilla, M.A, Martin-Mora, D, Krell, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
4HDQ
 
 | |
1FMO
 
 | | CRYSTAL STRUCTURE OF A POLYHISTIDINE-TAGGED RECOMBINANT CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH THE PEPTIDE INHIBITOR PKI(5-24) AND ADENOSINE | | Descriptor: | ADENOSINE, CAMP-DEPENDENT PROTEIN KINASE, HEAT STABLE RABBIT SKELETAL MUSCLE INHIBITOR PROTEIN | | Authors: | Narayana, N, Cox, S, Shaltiel, S, Taylor, S.S, Xuong, N.-H. | | Deposit date: | 1997-07-08 | | Release date: | 1998-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a polyhistidine-tagged recombinant catalytic subunit of cAMP-dependent protein kinase complexed with the peptide inhibitor PKI(5-24) and adenosine.
Biochemistry, 36, 1997
|
|
1DFK
 
 | |
1EMU
 
 | |
4HDO
 
 | | Crystal structure of the binary Complex of KRIT1 bound to the Rap1 GTPase | | Descriptor: | GLYCEROL, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Gingras, A.R. | | Deposit date: | 2012-10-02 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Structure of the Ternary Complex of Krev Interaction Trapped 1 (KRIT1) Bound to Both the Rap1 GTPase and the Heart of Glass (HEG1) Cytoplasmic Tail.
J.Biol.Chem., 288, 2013
|
|
1DEB
 
 | |
