8QMR
 
 | |
8QMT
 
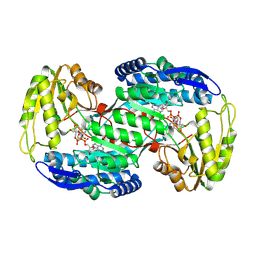 | | Succinic semialdehyde dehydrogenase from E. coli with Q262R substitution and bound NAD+, succinic semialdehyde | | Descriptor: | 4-oxobutanoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Succinate semialdehyde dehydrogenase [NAD(P)+] Sad | | Authors: | He, H, Zarzycki, J, Erb, T.J. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Adaptive laboratory evolution recruits the promiscuity of succinate semialdehyde dehydrogenase to repair different metabolic deficiencies.
Nat Commun, 15, 2024
|
|
8QMS
 
 | |
8QCZ
 
 | |
5C1C
 
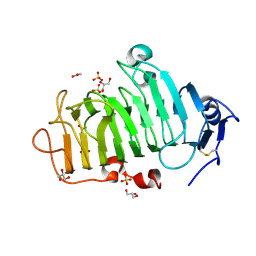 | | Crystal Structure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jameson, G.B, Williams, M.A.K, Loo, T.S, Kent, L.M, Melton, L.D, Mercadante, D. | | Deposit date: | 2015-06-13 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Properties of a Non-processive, Salt-requiring, and Acidophilic Pectin Methylesterase from Aspergillus niger Provide Insights into the Key Determinants of Processivity Control.
J.Biol.Chem., 291, 2016
|
|
5C1X
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound VIII | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
8Q6T
 
 | |
8QCB
 
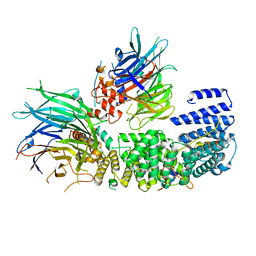 | | CryoEM structure of a S. Cerevisiae Ski2387 complex in the open state | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, Superkiller protein 3, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
5BZH
 
 | |
3FYC
 
 | |
3GP8
 
 | | Crystal structure of the binary complex of RecD2 with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*T*TP*TP*TP*TP*TP*TP*TP*T)-3', Exodeoxyribonuclease V, subunit RecD, ... | | Authors: | Saikrishnan, K, Cook, N, Wigley, D.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic basis of 5'-3' translocation in SF1B helicases.
Cell(Cambridge,Mass.), 137, 2009
|
|
5A6J
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
3GNU
 
 | | Toxin fold as basis for microbial attack and plant defense | | Descriptor: | 25 kDa protein elicitor, CHLORIDE ION, GUANIDINE | | Authors: | Ottmann, C, Luberacki, B, Kuefner, I, Koch, W, Brunner, F, Weyand, M, Mattinen, L, Pirhonen, M, Anderluh, G, Seitz, H.U, Nuernberger, T, Oecking, C. | | Deposit date: | 2009-03-18 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A common toxin fold mediates microbial attack and plant defense
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5B05
 
 | | Lysozyme (control experiment) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
5CMX
 
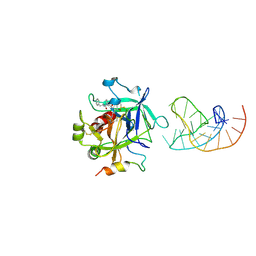 | | X-ray structure of the complex between human alpha thrombin and a duplex/quadruplex 31-mer DNA aptamer | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, RE31, ... | | Authors: | Russo Krauss, I, Pica, A, Napolitano, V, Sica, F. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding.
Nucleic Acids Res., 44, 2016
|
|
3GWG
 
 | |
3GJP
 
 | |
5B1Z
 
 | |
5B59
 
 | | Hen egg-white lysozyme modified with a keto-ABNO. | | Descriptor: | (1~{S},5~{R})-9-oxidanyl-9-azabicyclo[3.3.1]nonan-3-one, Lysozyme C | | Authors: | Sasaki, D, Seki, Y, Sohma, Y, Oisaki, K, Kanai, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transition Metal-Free Tryptophan-Selective Bioconjugation of Proteins
J.Am.Chem.Soc., 138, 2016
|
|
5B06
 
 | | Lysozyme (denatured by NaOD and refolded) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
5CK7
 
 | |
3GRU
 
 | |
3K3U
 
 | |
3H1F
 
 | |
3H3P
 
 | | Crystal structure of HIV epitope-scaffold 4E10 Fv complex | | Descriptor: | 4E10_S0_1TJLC_004_N, CALCIUM ION, Fv 4E10 heavy chain, ... | | Authors: | Holmes, M.A. | | Deposit date: | 2009-04-16 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between lipids and human anti-HIV antibody 4E10 can be reduced without ablating neutralizing activity
J.Virol., 84, 2010
|
|
