1L41
 
 | |
1L48
 
 | |
1L51
 
 | |
2ZNF
 
 | | HIGH-RESOLUTION STRUCTURE OF AN HIV ZINC FINGERLIKE DOMAIN VIA A NEW NMR-BASED DISTANCE GEOMETRY APPROACH | | Descriptor: | GAG POLYPROTEIN, ZINC ION | | Authors: | Summers, M.F, South, T.L, Kim, B, Hare, D.R. | | Deposit date: | 1990-03-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach.
Biochemistry, 29, 1990
|
|
1L60
 
 | |
1L67
 
 | |
1L74
 
 | |
1L37
 
 | |
1L59
 
 | |
1L56
 
 | |
1L70
 
 | |
1L75
 
 | |
2GCT
 
 | |
6ABP
 
 | |
3RN3
 
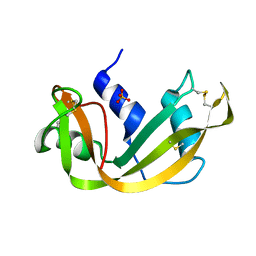 | | SEGMENTED ANISOTROPIC REFINEMENT OF BOVINE RIBONUCLEASE A BY THE APPLICATION OF THE RIGID-BODY TLS MODEL | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Howlin, B, Moss, D.S, Harris, G.W, Palmer, R.A. | | Deposit date: | 1991-10-30 | | Release date: | 1991-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Segmented anisotropic refinement of bovine ribonuclease A by the application of the rigid-body TLS model.
Acta Crystallogr.,Sect.A, 45, 1989
|
|
1LMB
 
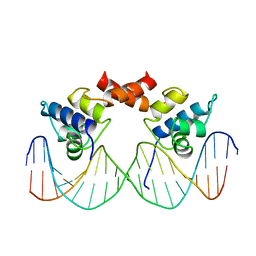 | | REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Beamer, L.J, Pabo, C.O. | | Deposit date: | 1991-11-05 | | Release date: | 1991-11-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A crystal structure of the lambda repressor-operator complex.
J.Mol.Biol., 227, 1992
|
|
9INS
 
 | |
4CMS
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES IV. STRUCTURE AND REFINEMENT AT 2.2 ANGSTROMS RESOLUTION OF BOVINE CHYMOSIN | | Descriptor: | CHYMOSIN B | | Authors: | Newman, M, Frazao, C, Khan, G, Tickle, I.J, Blundell, T.L, Safro, M, Andreeva, N, Zdanov, A. | | Deposit date: | 1991-11-01 | | Release date: | 1991-11-07 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray analyses of aspartic proteinases. IV. Structure and refinement at 2.2 A resolution of bovine chymosin.
J.Mol.Biol., 221, 1991
|
|
2BPA
 
 | | ATOMIC STRUCTURE OF SINGLE-STRANDED DNA BACTERIOPHAGE PHIX174 AND ITS FUNCTIONAL IMPLICATIONS | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*C)-3'), PROTEIN (SUBUNIT OF BACTERIOPHAGE PHIX174) | | Authors: | McKenna, R, Xia, D, Willingmann, P, Ilag, L.L, Krishnaswamy, S, Rossmann, M.G, Olson, N.H, Baker, T.S, Incardona, N.L. | | Deposit date: | 1991-12-03 | | Release date: | 1991-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Atomic structure of single-stranded DNA bacteriophage phi X174 and its functional implications.
Nature, 355, 1992
|
|
1PAL
 
 | | IONIC INTERACTIONS WITH PARVALBUMINS. CRYSTAL STRUCTURE DETERMINATION OF PIKE 4.10 PARVALBUMIN IN FOUR DIFFERENT IONIC ENVIRONMENTS | | Descriptor: | AMMONIUM ION, CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J, Rambaud, J. | | Deposit date: | 1990-11-08 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ionic interactions with parvalbumins. Crystal structure determination of pike 4.10 parvalbumin in four different ionic environments.
J.Mol.Biol., 220, 1991
|
|
5P21
 
 | |
1C2R
 
 | | MOLECULAR STRUCTURE OF CYTOCHROME C2 ISOLATED FROM RHODOBACTER CAPSULATUS DETERMINED AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Benning, M.M, Wesenberg, G, Caffrey, M.S, Bartsch, R.G, Meyer, T.E, Cusanovich, M.A, Rayment, I, Holden, H.M. | | Deposit date: | 1991-03-19 | | Release date: | 1992-01-15 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular structure of cytochrome c2 isolated from Rhodobacter capsulatus determined at 2.5 A resolution.
J.Mol.Biol., 220, 1991
|
|
1MSB
 
 | |
1HDD
 
 | | CRYSTAL STRUCTURE OF AN ENGRAILED HOMEODOMAIN-DNA COMPLEX AT 2.8 ANGSTROMS RESOLUTION: A FRAMEWORK FOR UNDERSTANDING HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*G P*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*C P*CP*TP*AP*A)-3'), PROTEIN (ENGRAILED HOMEODOMAIN) | | Authors: | Kissinger, C.R, Liu, B, Martin-Blanco, E, Kornberg, T.B, Pabo, C.O. | | Deposit date: | 1991-09-16 | | Release date: | 1992-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 63, 1990
|
|
1MVP
 
 | |
