7JHU
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J33 immobile Holliday junction with R3 symmetry | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*CP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JK0
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J1 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*TP*GP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.058 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JIO
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J23 immobile Holliday junction with R3 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*GP*GP*TP*GP*TP*CP*GP*T)-3'), DNA (5'-D(P*CP*GP*AP*CP*GP*AP*CP*TP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JJ5
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J10 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*GP*GP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-24 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JJX
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J5 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*CP*GP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JFT
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J2 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*CP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.158 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JH9
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J22 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*GP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.094 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JHR
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J4 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*CP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.158 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JJ4
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J14 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*GP*CP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-24 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.022 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
1R5S
 
 | | Connexin 43 Carboxyl Terminal Domain | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Sorgen, P.L, Duffy, H.S, Mario, D, Sahoo, P, Coombs, W, Delmar, M, Spray, D.C. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1
J.Biol.Chem., 279, 2004
|
|
7K3P
 
 | |
7ESS
 
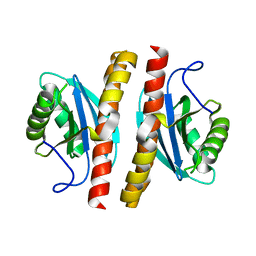 | | Structure-guided studies of the Holliday junction resolvase RuvX provide novel insights into ATP-stimulated cleavage of branched DNA and RNA substrates | | Descriptor: | Putative pre-16S rRNA nuclease | | Authors: | Thakur, M, Mohan, D, Singh, A.K, Agarwal, A, Gopal, B, Muniyappa, K. | | Deposit date: | 2021-05-11 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Novel insights into ATP-Stimulated Cleavage of branched DNA and RNA Substrates through Structure-Guided Studies of the Holliday Junction Resolvase RuvX.
J.Mol.Biol., 433, 2021
|
|
2X61
 
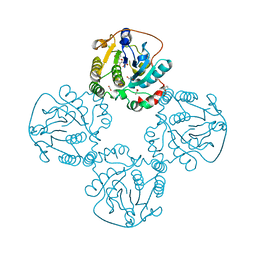 | | Crystal structure of the sialyltransferase CST-II in complex with trisaccharide acceptor and CMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
8GH8
 
 | | RuvA Holliday junction DNA complex | | Descriptor: | DNA (34-MER), Holliday junction branch migration complex subunit RuvA | | Authors: | Rish, A.D, Fu, T. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | RuvA Holliday junction DNA complex
To Be Published
|
|
6GDS
 
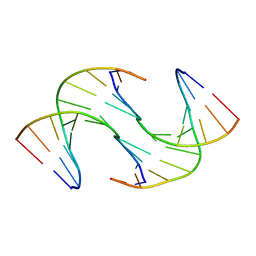 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | Telomeric DNA (5' CTAACCCTAA) 10mer, Telomeric DNA (5'-TTAGGGTTAG)-3') 10mer | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
1RK8
 
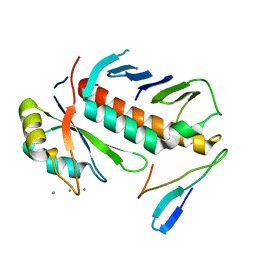 | | Structure of the cytosolic protein PYM bound to the Mago-Y14 core of the exon junction complex | | Descriptor: | CALCIUM ION, CG8781-PA protein, Mago nashi protein, ... | | Authors: | Bono, F, Ebert, J, Guettler, T, Izaurralde, E, Conti, E. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the interaction of PYM with
the Mago-Y14 core of the exon junction complex
EMBO Rep., 5, 2004
|
|
8SH1
 
 | |
8SH0
 
 | |
2X63
 
 | | Crystal structure of the sialyltransferase CST-II N51A in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
2X62
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE CST-II Y81F IN COMPLEX WITH CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, N3-PROTONATED CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
6GDH
 
 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*AP*CP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3') | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
6GDN
 
 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | MAGNESIUM ION, Telomere DNA (42-MER) | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
1SQ1
 
 | | Crystal Structure of the Chorismate Synthase from Campylobacter jejuni, Northeast Structural Genomics Target BR19 | | Descriptor: | Chorismate synthase, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Chorismate Synthase from Campylobacter jejuni, Northeast Structural Genomics Target BR19
To be Published
|
|
7JLF
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J24 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*AP*GP*AP*CP*GP*TP*CP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKE
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J2 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*AP*CP*GP*AP*GP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.068 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
