1FI9
 
 | | SOLUTION STRUCTURE OF THE IMIDAZOLE COMPLEX OF CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1FIA
 
 | | CRYSTAL STRUCTURE OF THE FACTOR FOR INVERSION STIMULATION FIS AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Kostrewa, D, Granzin, J, Choe, H.-W, Labahn, J, Saenger, W. | | Deposit date: | 1991-12-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the factor for inversion stimulation FIS at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
1FIB
 
 | |
1FIC
 
 | |
1FID
 
 | |
1FIE
 
 | | RECOMBINANT HUMAN COAGULATION FACTOR XIII | | Descriptor: | COAGULATION FACTOR XIII | | Authors: | Yee, V.C, Teller, D.C. | | Deposit date: | 1996-08-24 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural evidence that the activation peptide is not released upon thrombin cleavage of factor XIII.
Thromb.Res., 78, 1995
|
|
1FIF
 
 | | N-ACETYLGALACTOSAMINE-SELECTIVE MUTANT OF MANNOSE-BINDING PROTEIN-A (QPDWG-HDRPY) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-A | | Authors: | Feinberg, H, Torgersen, D, Drickamer, K, Weis, W.I. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of pH-dependent N-acetylgalactosamine binding by a functional mimic of the hepatocyte asialoglycoprotein receptor.
J.Biol.Chem., 275, 2000
|
|
1FIG
 
 | | ROUTES TO CATALYSIS: STRUCTURE OF A CATALYTIC ANTIBODY AND COMPARISON WITH ITS NATURAL COUNTERPART | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, IGG1-KAPPA 1F7 FAB (HEAVY CHAIN), IGG1-KAPPA 1F7 FAB (LIGHT CHAIN) | | Authors: | Haynes, M.R, Stura, E.A, Hilvert, D, Wilson, I.A. | | Deposit date: | 1994-01-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Routes to catalysis: structure of a catalytic antibody and comparison with its natural counterpart.
Science, 263, 1994
|
|
1FIH
 
 | | N-ACETYLGALACTOSAMINE BINDING MUTANT OF MANNOSE-BINDING PROTEIN A (QPDWG-HDRPY), COMPLEX WITH N-ACETYLGALACTOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Feinberg, H, Torgerson, D, Drickamer, K, Weis, W.I. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of pH-dependent N-acetylgalactosamine binding by a functional mimic of the hepatocyte asialoglycoprotein receptor.
J.Biol.Chem., 275, 2000
|
|
1FIK
 
 | |
1FIL
 
 | |
1FIM
 
 | |
1FIN
 
 | |
1FIO
 
 | | CRYSTAL STRUCTURE OF YEAST T-SNARE PROTEIN SSO1 | | Descriptor: | SSO1 PROTEIN, ZINC ION | | Authors: | Munson, M, Chen, X, Cocina, A.E, Schultz, S.M, Hughson, F.M. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly.
Nat.Struct.Biol., 7, 2000
|
|
1FIP
 
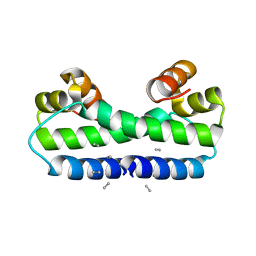 | | THE STRUCTURE OF FIS MUTANT PRO61ALA ILLUSTRATES THAT THE KINK WITHIN THE LONG ALPHA-HELIX IS NOT DUE TO THE PRESENCE OF THE PROLINE RESIDUE | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS), UNKNOWN PEPTIDE, POSSIBLY PART OF THE UNOBSERVED RESIDUES IN ENTITY 1 | | Authors: | Yuan, H.S, Wang, S.S, Yang, W.-Z, Finkel, S.E, Johnson, R.C. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of Fis mutant Pro61Ala illustrates that the kink within the long alpha-helix is not due to the presence of the proline residue.
J.Biol.Chem., 269, 1994
|
|
1FIQ
 
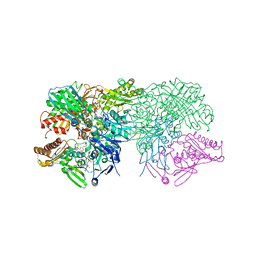 | | CRYSTAL STRUCTURE OF XANTHINE OXIDASE FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FIR
 
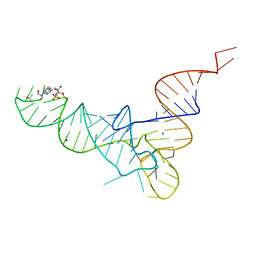 | |
1FIT
 
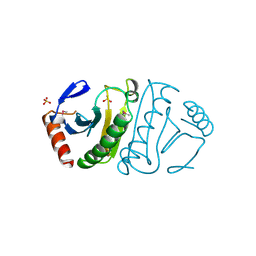 | | FHIT (FRAGILE HISTIDINE TRIAD PROTEIN) | | Descriptor: | FRAGILE HISTIDINE PROTEIN, SULFATE ION, beta-D-fructofuranose | | Authors: | Lima, C.D, D'Amico, K.L, Naday, I, Rosenbaum, G, Westbrook, E.M, Hendrickson, W.A. | | Deposit date: | 1997-05-17 | | Release date: | 1997-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family.
Structure, 5, 1997
|
|
1FIU
 
 | | TETRAMERIC RESTRICTION ENDONUCLEASE NGOMIV IN COMPLEX WITH CLEAVED DNA | | Descriptor: | ACETIC ACID, DNA (5'-D(*TP*GP*CP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*CP*GP*C)-3'), ... | | Authors: | Deibert, M, Grazulis, S, Sasnauskas, G, Siksnys, V, Huber, R. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA.
Nat.Struct.Biol., 7, 2000
|
|
1FIV
 
 | | STRUCTURE OF AN INHIBITOR COMPLEX OF PROTEINASE FROM FELINE IMMUNODEFICIENCY VIRUS | | Descriptor: | FIV PROTEASE, FIV PROTEASE INHIBITOR ACE-ALN-VAL-STA-GLU-ALN-NH2 | | Authors: | Wlodawer, A, Gustchina, A, Reshetnikova, L, Lubkowski, J, Zdanov, A. | | Deposit date: | 1995-05-04 | | Release date: | 1995-07-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an inhibitor complex of the proteinase from feline immunodeficiency virus.
Nat.Struct.Biol., 2, 1995
|
|
1FIW
 
 | | THREE-DIMENSIONAL STRUCTURE OF BETA-ACROSIN FROM RAM SPERMATOZOA | | Descriptor: | BETA-ACROSIN HEAVY CHAIN, BETA-ACROSIN LIGHT CHAIN, P-AMINO BENZAMIDINE, ... | | Authors: | Tranter, R, Read, J.A, Jones, R, Brady, R.L. | | Deposit date: | 2000-08-07 | | Release date: | 2000-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effector sites in the three-dimensional structure of mammalian sperm beta-acrosin.
Structure Fold.Des., 8, 2000
|
|
1FIX
 
 | |
1FIY
 
 | | THREE-DIMENSIONAL STRUCTURE OF PHOSPHOENOLPYRUVATE CARBOXYLASE FROM ESCHERICHIA COLI AT 2.8 A RESOLUTION | | Descriptor: | ASPARTIC ACID, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Kai, Y, Matsumura, H, Inoue, T, Terada, K, Nagara, Y, Yoshinaga, T, Kihara, A, Izui, K. | | Deposit date: | 1998-05-02 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1FIZ
 
 | | THREE DIMENSIONAL STRUCTURE OF BETA-ACROSIN FROM BOAR SPERMATOZOA | | Descriptor: | BETA-ACROSIN HEAVY CHAIN, BETA-ACROSIN LIGHT CHAIN, P-AMINO BENZAMIDINE, ... | | Authors: | Tranter, R, Read, J.A, Jones, R, Brady, R.L. | | Deposit date: | 2000-08-07 | | Release date: | 2000-11-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Effector sites in the three-dimensional structure of mammalian sperm beta-acrosin.
Structure Fold.Des., 8, 2000
|
|
1FJ0
 
 | |
