6J26
 
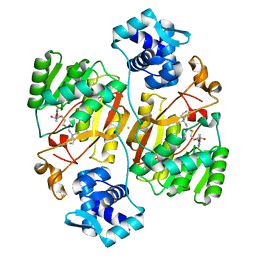 | | Crystal structure of the branched-chain polyamine synthase from Thermococcus kodakarensis (Tk-BpsA) in complex with N4-bis(aminopropyl)spermidine and 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, FE (III) ION, N(4)-bis(aminopropyl)spermidine synthase, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
3EPH
 
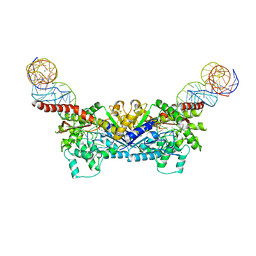 | |
6J40
 
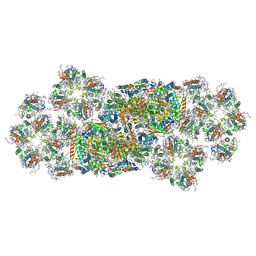 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
6JLJ
 
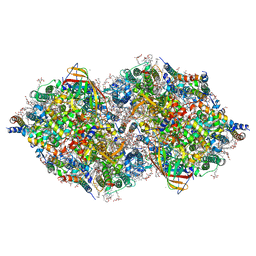 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JO3
 
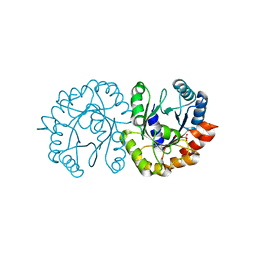 | | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with substrate sn-glycerol-1-phosphate | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE | | Authors: | Nemoto, N, Miyazono, K, Tanokura, M, Yamagishi, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with the substrate sn-glycerol 1-phosphate.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6JLM
 
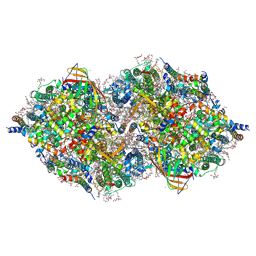 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JCN
 
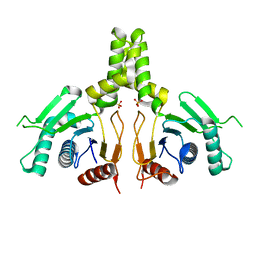 | | Yeast dehydrodolichyl diphosphate synthase complex subunit NUS1 | | Descriptor: | Dehydrodolichyl diphosphate synthase complex subunit NUS1, SULFATE ION | | Authors: | Ko, T.-P, Ma, J, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-01-29 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural insights to heterodimeric cis-prenyltransferases through yeast dehydrodolichyl diphosphate synthase subunit Nus1.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
3GKF
 
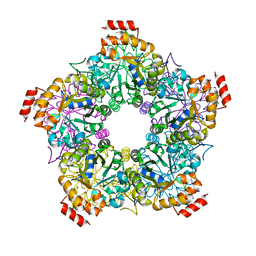 | | Crystal Structure of E. coli LsrF | | Descriptor: | Aldolase lsrF | | Authors: | Miller, S.T, Diaz, Z.C. | | Deposit date: | 2009-03-10 | | Release date: | 2009-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the Escherichia coli autoinducer-2 processing protein LsrF.
Plos One, 4, 2009
|
|
3GLC
 
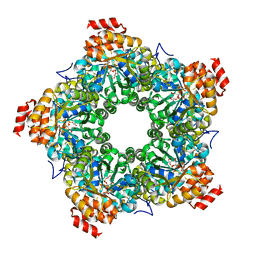 | |
3GOS
 
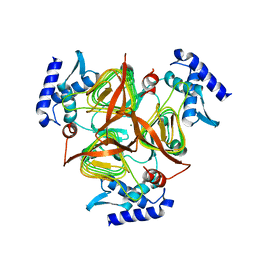 | | The crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Yersinia pestis CO92 | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase, MAGNESIUM ION | | Authors: | Zhang, R, Maltseva, N, Kwon, K, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-19 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Yersinia pestis CO92
To be Published
|
|
3H9K
 
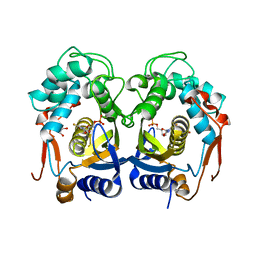 | | Structures of Thymidylate Synthase R163K with Substrates and Inhibitors Show Subunit Asymmetry | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Gibson, L.M, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2009-04-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of human thymidylate synthase R163K with dUMP, FdUMP and glutathione show asymmetric ligand binding.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3H49
 
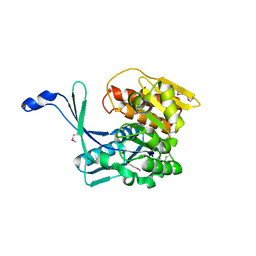 | |
3EUQ
 
 | |
3EUT
 
 | |
3EZE
 
 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | Descriptor: | PHOSPHITE ION, PROTEIN (PHOSPHOTRANSFERASE SYSTEM, ENZYME I), ... | | Authors: | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1998-11-04 | | Release date: | 1998-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
3EY5
 
 | | Putative acetyltransferase from GNAT family from Bacteroides thetaiotaomicron. | | Descriptor: | Acetyltransferase-like, GNAT family, SULFATE ION | | Authors: | Osipiuk, J, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-17 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystal structure of putative acetyltransferase from GNAT family from Bacteroides thetaiotaomicron.
To be Published
|
|
3F0A
 
 | | Structure of a putative n-acetyltransferase (ta0374) in complex with acetyl-coa from thermoplasma acidophilum | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, N-ACETYLTRANSFERASE, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3HBB
 
 | | Structures of dihydrofolate reductase-thymidylate synthase of Trypanosoma cruzi in the folate-free state and in complex with two antifolate drugs, trimetrexate and methotrexate | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of dihydrofolate reductase-thymidylate synthase of Trypanosoma cruzi in the folate-free state and in complex with two antifolate drugs, trimetrexate and methotrexate.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3GYA
 
 | |
3EN9
 
 | | Structure of the Methanococcus jannaschii KAE1-BUD32 fusion protein | | Descriptor: | HEXATANTALUM DODECABROMIDE, MAGNESIUM ION, O-sialoglycoprotein endopeptidase/protein kinase | | Authors: | Neculai, D. | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
3EPJ
 
 | |
3ELU
 
 | | Wesselsbron virus Methyltransferase in complex with AdoMet | | Descriptor: | Methyltransferase, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Bollati, M, Ricagno, S, Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
3EMD
 
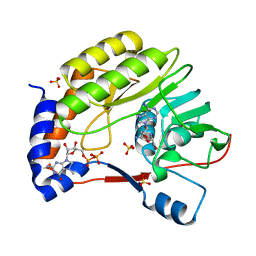 | | Wesselsbron virus Methyltransferase in complex with Sinefungin and 7MeGpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, SINEFUNGIN, SULFATE ION, ... | | Authors: | Bollati, M, Milani, M, Ricagno, S, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-24 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
3EPK
 
 | |
3GND
 
 | |
