7P7S
 
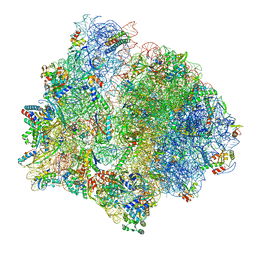 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state II | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7U
 
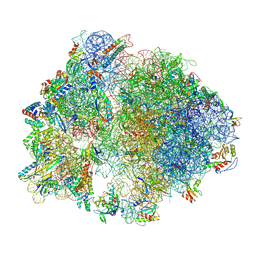 | | E. faecalis 70S ribosome with P-tRNA, state IV | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7Q
 
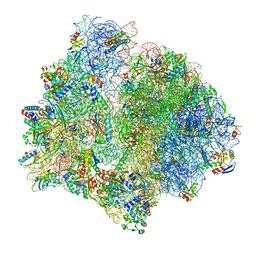 | | E. faecalis 70S ribosome bound by PoxtA-EQ2, high-resolution combined volume | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7R
 
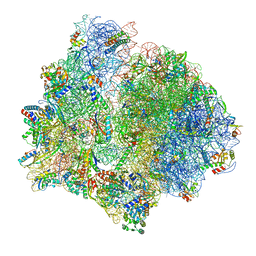 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state I | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7Q37
 
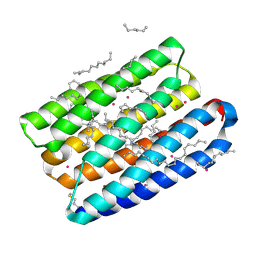 | | Crystal structure of proton pump MAR rhodopsin pressurized with krypton | | Descriptor: | Bacteriorhodopsin, EICOSANE, HEXANE, ... | | Authors: | Melnikov, I, Rulev, M, Astashkin, R, Kovalev, K, Carpentier, P, Gordeliy, V, Popov, A. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-pressure crystallography shows noble gas intervention into protein-lipid interaction and suggests a model for anaesthetic action.
Commun Biol, 5, 2022
|
|
5O0S
 
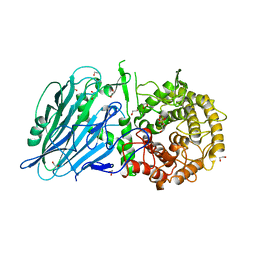 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with unreacted beta Cyclophellitol Cyclosulfate probe ME711 | | Descriptor: | (3~{a}~{S},4~{R},5~{S},6~{R},7~{R},7~{a}~{R})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydrobenzo[d][1,3,2]dioxathiole-4,5,6-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
5OLE
 
 | |
5O4W
 
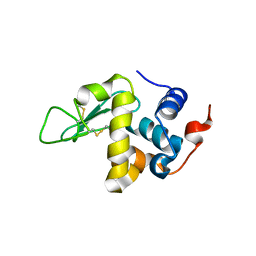 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | Descriptor: | Lysozyme C | | Authors: | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | Cite: | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5O4X
 
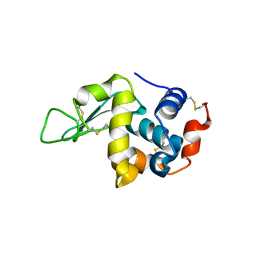 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | Descriptor: | Lysozyme C | | Authors: | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | Cite: | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NPF
 
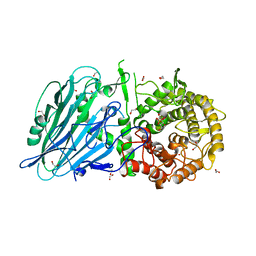 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with beta Cyclophellitol Cyclosulfate probe ME594 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucosylceramidase, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
7O6N
 
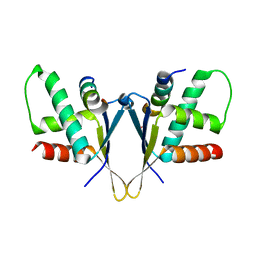 | | Crystal structure of C. elegans ERH-2 PID-3 complex | | Descriptor: | Enhancer of rudimentary homolog 2, FORMIC ACID, Protein pid-3 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
7OCX
 
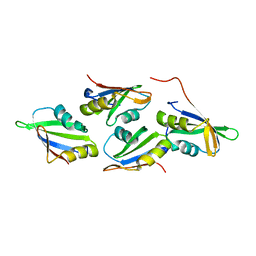 | |
7OCZ
 
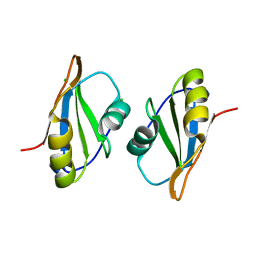 | |
7O6L
 
 | | Crystal structure of C. elegans ERH-2 | | Descriptor: | Enhancer of rudimentary homolog 2 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
7O7K
 
 | | Crystal structure of the human DYRK1A kinase domain bound to abemaciclib | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaltheuner, I.H, Anand, K, Geyer, M. | | Deposit date: | 2021-04-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation.
Nat Commun, 12, 2021
|
|
4H4A
 
 | |
8EUA
 
 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | Descriptor: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | Authors: | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | Deposit date: | 2022-10-18 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
5XYW
 
 | |
5FIQ
 
 | | Exonuclease domain-containing 1 (Exd1) in the native conformation | | Descriptor: | EXD1 | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
5XYV
 
 | |
5FIS
 
 | | Exonuclease domain-containing 1 (Exd1) in the Gd bound conformation | | Descriptor: | EXD1, GADOLINIUM ATOM | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-02 | | Release date: | 2015-12-23 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
8DL9
 
 | |
8DLB
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease in complex with compound Z2799209083 | | Descriptor: | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase | | Authors: | Kovalevsky, A.Y, Coates, L, Kneller, D.W. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AI-Accelerated Design of Targeted Covalent Inhibitors for SARS-CoV-2.
J.Chem.Inf.Model., 63, 2023
|
|
8DMD
 
 | |
6AKL
 
 | | Crystal structure of Striatin3 in complex with SIKE1 Coiled-coil domain | | Descriptor: | Striatin-3, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
