1F9P
 
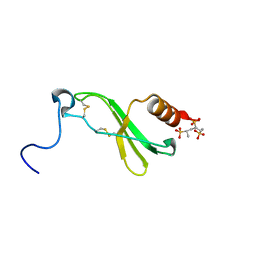 | | CRYSTAL STRUCTURE OF CONNECTIVE TISSUE ACTIVATING PEPTIDE-III(CTAP-III) COMPLEXED WITH POLYVINYLSULFONIC ACID | | Descriptor: | CONNECTIVE TISSUE ACTIVATING PEPTIDE-III, ETHANESULFONIC ACID | | Authors: | Yang, J, Faulk, T, Aster, R, Visentin, G, Edwards, B, Castor, C. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the CXC Chemokine, Connective Tissue Activating Peptide-III, Complexed with the Heparin Analogue, Polyvinylsulfonic Acid
To be Published
|
|
1F9Q
 
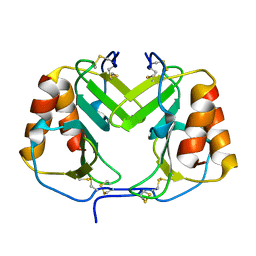 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-Type Reveals the Epitopes for the Heparin-Induced Thrombocytopenia Antibodies
To be Published
|
|
1F9R
 
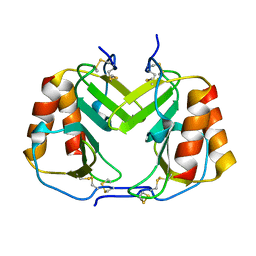 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 MUTANT 1 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-type Reveals the Epitopes for the Heparin-induced Thrombocytopenia Antibodies
To be Published
|
|
1F9S
 
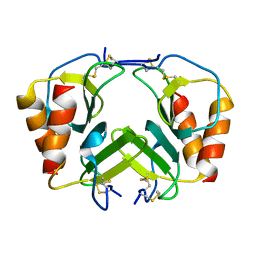 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 MUTANT 2 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-type Reveals the Epitopes for the Heparin-induced Thrombocytopenia Antibodies
To be Published
|
|
1F9T
 
 | | CRYSTAL STRUCTURES OF KINESIN MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE MOTOR ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Yun, M, Zhang, X, Park, C.-G, Park, H.-W, Endow, S.A. | | Deposit date: | 2000-07-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1F9U
 
 | | CRYSTAL STRUCTURES OF MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE KINESIN MOTOR ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Yun, M, Zhang, X, Park, C.G, Park, H.W, Endow, S.A. | | Deposit date: | 2000-07-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1F9V
 
 | | CRYSTAL STRUCTURES OF MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE KINESIN MOTOR ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Yun, M, Zhang, X, Park, C.G, Park, H.W, Endow, S.A. | | Deposit date: | 2000-07-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1F9W
 
 | | CRYSTAL STRUCTURES OF MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE KINESIN MOTOR ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Yun, M, Zhang, X, Park, C.G, Park, H.W, Endow, S.A. | | Deposit date: | 2000-07-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1F9X
 
 | | AVERAGE NMR SOLUTION STRUCTURE OF THE BIR-3 DOMAIN OF XIAP | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN XIAP, ZINC ION | | Authors: | Sun, C, Cai, M, Meadows, R.P, Fesik, S.W. | | Deposit date: | 2000-07-11 | | Release date: | 2001-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP.
J.Biol.Chem., 275, 2000
|
|
1F9Z
 
 | | CRYSTAL STRUCTURE OF THE NI(II)-BOUND GLYOXALASE I FROM ESCHERICHIA COLI | | Descriptor: | GLYOXALASE I, NICKEL (II) ION | | Authors: | He, M.M, Clugston, S.L, Honek, J.F, Matthews, B.W. | | Deposit date: | 2000-07-11 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation.
Biochemistry, 39, 2000
|
|
1FA0
 
 | | STRUCTURE OF YEAST POLY(A) POLYMERASE BOUND TO MANGANATE AND 3'-DATP | | Descriptor: | 3'-DEOXYADENOSINE, 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Bard, J, Zhelkovsky, A.M, Helmling, S, Moore, C.L, Bohm, A. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of yeast poly(A) polymerase alone and in complex with 3'-dATP.
Science, 289, 2000
|
|
1FA2
 
 | | CRYSTAL STRUCTURE OF BETA-AMYLASE FROM SWEET POTATO | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-AMYLASE, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Lee, B.I, Cheong, C.G, Suh, S.W. | | Deposit date: | 2000-07-12 | | Release date: | 2000-08-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization, molecular replacement solution, and refinement of tetrameric beta-amylase from sweet potato.
Proteins, 21, 1995
|
|
1FA3
 
 | |
1FA4
 
 | | ELUCIDATION OF THE PARAMAGNETIC RELAXATION OF HETERONUCLEI AND PROTONS IN CU(II) PLASTOCYANIN FROM ANABAENA VARIABILIS | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Ma, L, Jorgensen, A.M, Sorensen, G.O, Ulstrup, J, Led, J.J. | | Deposit date: | 2000-07-12 | | Release date: | 2000-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Elucidation of the Paramagnetic R1 Relaxation of Heteronuclei and Protons in Cu(II) Plastocyanin from Anabaena Variabilis
J.Am.Chem.Soc., 122, 2000
|
|
1FA5
 
 | | CRYSTAL STRUCTURE OF THE ZN(II)-BOUND GLYOXALASE I OF ESCHERICHIA COLI | | Descriptor: | GLYOXALASE I, ZINC ION | | Authors: | He, M.M, Clugston, S.L, Honek, J.F, Matthews, B.W. | | Deposit date: | 2000-07-12 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation.
Biochemistry, 39, 2000
|
|
1FA6
 
 | | CRYSTAL STRUCTURE OF THE CO(II)-BOUND GLYOXALASE I OF ESCHERICHIA COLI | | Descriptor: | COBALT (II) ION, GLYOXALASE I | | Authors: | He, M.M, Clugston, S.L, Honek, J.F, Matthews, B.W. | | Deposit date: | 2000-07-12 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation.
Biochemistry, 39, 2000
|
|
1FA7
 
 | | CRYSTAL STRUCTURE OF CD(II)-BOUND GLYOXALASE I OF ESCHERICHIA COLI | | Descriptor: | CADMIUM ION, GLYOXALASE I | | Authors: | He, M.M, Clugston, S.L, Honek, J.F, Matthews, B.W. | | Deposit date: | 2000-07-12 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation.
Biochemistry, 39, 2000
|
|
1FA8
 
 | |
1FA9
 
 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCOGEN PHOSPHORYLASE, LIVER FORM, ... | | Authors: | Rath, V.L, Ammirati, M, LeMotte, P.K, Fennell, K.F, Mansour, M.N, Danley, D.E, Hynes, T.R, Schulte, G.K, Wasilko, D.J, Pandit, J. | | Deposit date: | 2000-07-12 | | Release date: | 2000-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core.
Mol.Cell, 6, 2000
|
|
1FAA
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN F FROM SPINACH CHLOROPLAST (LONG FORM) | | Descriptor: | THIOREDOXIN F | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-13 | | Release date: | 2000-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1FAC
 
 | |
1FAD
 
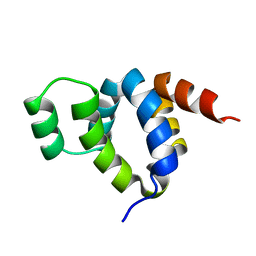 | | DEATH DOMAIN OF FAS-ASSOCIATED DEATH DOMAIN PROTEIN, RESIDUES 89-183 | | Descriptor: | PROTEIN (FADD PROTEIN) | | Authors: | Jeong, E.-J, Bang, S, Lee, T.H, Park, Y.-I, Sim, W.-S, Kim, K.-S. | | Deposit date: | 1999-03-23 | | Release date: | 1999-07-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD.
J.Biol.Chem., 274, 1999
|
|
1FAE
 
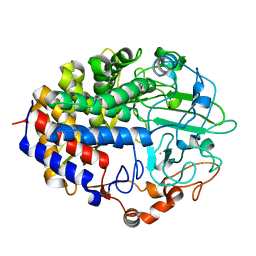 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-13 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1FAF
 
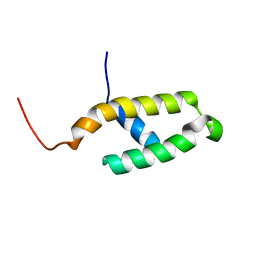 | | NMR STRUCTURE OF THE N-TERMINAL J DOMAIN OF MURINE POLYOMAVIRUS T ANTIGENS. | | Descriptor: | LARGE T ANTIGEN | | Authors: | Berjanskii, M.V, Riley, M.I, Xie, A, Semenchenko, V, Folk, W.R, Van Doren, S.R. | | Deposit date: | 2000-07-13 | | Release date: | 2000-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal J domain of murine polyomavirus T antigens. Implications for DnaJ-like domains and for mutations of T antigens.
J.Biol.Chem., 275, 2000
|
|
1FAG
 
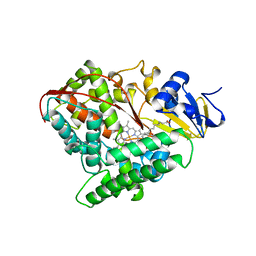 | | STRUCTURE OF CYTOCHROME P450 | | Descriptor: | CYTOCHROME P450 BM-3, PALMITOLEIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, H.Y, Poulos, T.L. | | Deposit date: | 1996-08-01 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid.
Nat.Struct.Biol., 4, 1997
|
|
