1HHH
 
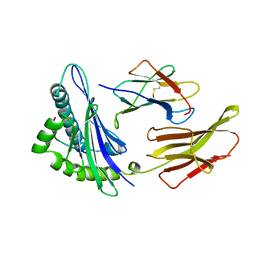 | | THE ANTIGENIC IDENTITY OF PEPTIDE(SLASH)MHC COMPLEXES: A COMPARISON OF THE CONFORMATION OF FIVE PEPTIDES PRESENTED BY HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), HEPATITIS B NUCLEOCAPSID PROTEIN (RESIDUES 18-27) | | Authors: | Madden, D.R, Garboczi, D.N, Wiley, D.C. | | Deposit date: | 1993-06-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2.
Cell(Cambridge,Mass.), 75, 1993
|
|
5SA9
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2697514548 | | Descriptor: | 1-methylindazole-3-carboxamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1424343998 | | Descriptor: | N-{2-[(propan-2-yl)sulfanyl]phenyl}urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA6
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2856434783 | | Descriptor: | N-benzyl-1-(4-fluorophenyl)methanamine, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.517 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z59181945 | | Descriptor: | CITRIC ACID, N-hydroxyquinoline-2-carboxamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1530301542 | | Descriptor: | 4-ethyl-2-(1H-imidazol-5-yl)-1,3-thiazole, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1673618163 | | Descriptor: | 4-amino-N-(2-hydroxyethyl)-N-methylbenzene-1-sulfonamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z31504642 | | Descriptor: | 2-methoxy-N-phenylacetamide, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAE
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z3219959731 | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease, pyridazin-3(2H)-one | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5E3O
 
 | |
1SZU
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
5SAG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-1605072 | | Descriptor: | 3-(1H-imidazol-2-yl)propan-1-amine, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAA
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z319891284 | | Descriptor: | 3-[(2S)-1-(methanesulfonyl)pyrrolidin-2-yl]-5-methyl-1,2-oxazole, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.239 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA4
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z239136710 | | Descriptor: | N-(5-methyl-1H-pyrazol-3-yl)acetamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAH
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-100112 | | Descriptor: | 2-methyl-5,6,7,8-tetrahydropyrido[4,3-c]pyridazin-3(2H)-one, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
1TAZ
 
 | | Catalytic Domain Of Human Phosphodiesterase 1B | | Descriptor: | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ZINC ION | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
3FHN
 
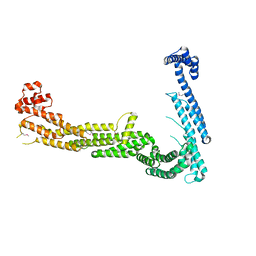 | | Structure of Tip20p | | Descriptor: | Protein transport protein TIP20 | | Authors: | Tripathi, A, Ren, Y, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5SA8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z68299550 | | Descriptor: | 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAD
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z425449682 | | Descriptor: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
3FKE
 
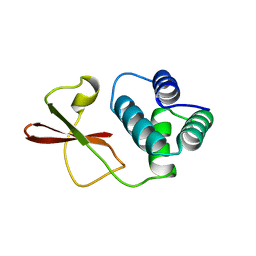 | | Structure of the Ebola VP35 Interferon Inhibitory Domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Amarasinghe, G.K, Leung, D.W, Ginder, N.D, Honzatko, R.B, Nix, J, Basler, C.F, Fulton, D.B. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Ebola VP35 interferon inhibitory domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZPU
 
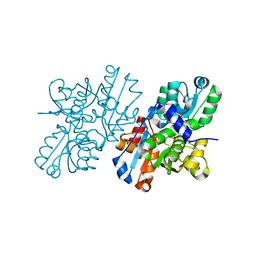 | | Crystal Structure of Modified Serine Racemase from S.pombe. | | Descriptor: | MAGNESIUM ION, N-(5'-PHOSPHOPYRIDOXYL)-D-ALANINE, Uncharacterized protein C320.14 | | Authors: | Goto, M. | | Deposit date: | 2008-07-29 | | Release date: | 2009-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serine racemase with catalytically active lysinoalanyl residue.
J.Biochem., 145, 2009
|
|
1D21
 
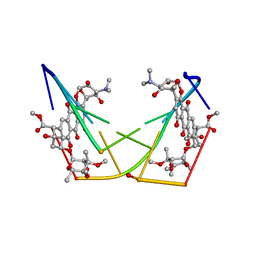 | | BINDING OF THE ANTITUMOR DRUG NOGALAMYCIN AND ITS DERIVATIVES TO DNA: STRUCTURAL COMPARISON | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*(AS)P*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Gao, Y.-G, Liaw, Y.-C, Robinson, H, Wang, A.H.-J. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of the antitumor drug nogalamycin and its derivatives to DNA: structural comparison.
Biochemistry, 29, 1990
|
|
3Q1O
 
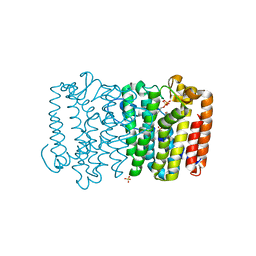 | | Crystal structure of geranyltransferase from helicobacter pylori complexed with magnesium and isoprenyl diphosphate | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Geranyltranstransferase (IspA), MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-17 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2ZBK
 
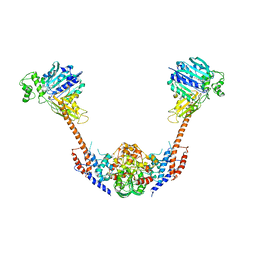 | | Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms | | Descriptor: | RADICICOL, Type 2 DNA topoisomerase 6 subunit B, Type II DNA topoisomerase VI subunit A | | Authors: | Graille, M, Cladiere, L, Durand, D, Lecointe, F, Forterre, P, van Tilbeurgh, H, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2007-10-22 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Crystal Structure of an Intact Type II DNA Topoisomerase: Insights into DNA Transfer Mechanisms
Structure, 16, 2008
|
|
4OB8
 
 | | Crystal structure of a novel thermostable esterase from Pseudomonas putida ECU1011 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
