7VYN
 
 | | Matrix arm of active state CI from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-14 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VZ8
 
 | | Membrane arm of deactive state CI from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VZ1
 
 | | Matrix arm of deactive state CI from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7WCP
 
 | |
7WCH
 
 | |
7W1P
 
 | | Deactive state CI from Q10-NADH dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2KPL
 
 | | MAGI-1 PDZ1 / E6CT | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, Protein E6 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
7W4M
 
 | | Deactive state CI from Q1-NADH dataset, Subclass 4 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4EDL
 
 | | Crystal structure of beta-parvin CH2 domain | | Descriptor: | 1,2-ETHANEDIOL, Beta-parvin | | Authors: | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
7W4L
 
 | | Deactive state CI from Q1-NADH dataset, Subclass 3 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W1Z
 
 | | Active state CI from Rotenone-NADH dataset, Subclass 2 | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-21 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4G
 
 | | Active state CI from Q1-NADH dataset, Subclass 5 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-27 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W2K
 
 | | Deactive state CI from Rotenone-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4F
 
 | | Active state CI from Q1-NADH dataset, Subclass 4 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-27 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4N
 
 | | Deactive state CI from Q1-NADH dataset, Subclass 5 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2KPK
 
 | | MAGI-1 PDZ1 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
4EDN
 
 | | Crystal structure of beta-parvin CH2 domain in complex with paxillin LD1 motif | | Descriptor: | Beta-parvin, Paxillin, SULFATE ION | | Authors: | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
1ATP
 
 | | 2.2 angstrom refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MNATP and a peptide inhibitor | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PEPTIDE INHIBITOR PKI(5-24), ... | | Authors: | Zheng, J, Trafny, E.A, Knighton, D.R, Xuong, N.-H, Taylor, S.S, Teneyck, L.F, Sowadski, J.M. | | Deposit date: | 1993-01-08 | | Release date: | 1993-04-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1C2O
 
 | | ELECTROPHORUS ELECTRICUS ACETYLCHOLINESTERASE | | Descriptor: | ACETYLCHOLINESTERASE | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Conformational flexibility of the acetylcholinesterase tetramer suggested by x-ray crystallography.
J.Biol.Chem., 274, 1999
|
|
192D
 
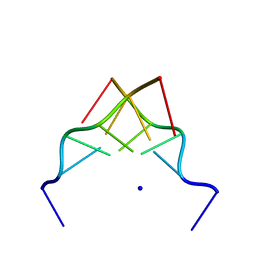 | | RECOMBINATION-LIKE STRUCTURE OF D(CCGCGG) | | Descriptor: | DNA (5'-D(*CP*CP*GP*CP*GP*G)-3'), SODIUM ION | | Authors: | Malinina, L, Urpi, L, Salas, X, Huynh-Dinh, T, Subirana, J.A. | | Deposit date: | 1994-09-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Recombination-like structure of d(CCGCGG).
J.Mol.Biol., 243, 1994
|
|
1BX6
 
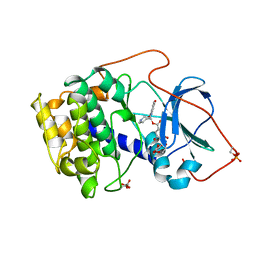 | | CRYSTAL STRUCTURE OF THE POTENT NATURAL PRODUCT INHIBITOR BALANOL IN COMPLEX WITH THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE | | Descriptor: | BALANOL, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Xuong, N.-H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1998-10-13 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase.
Biochemistry, 38, 1999
|
|
1BKX
 
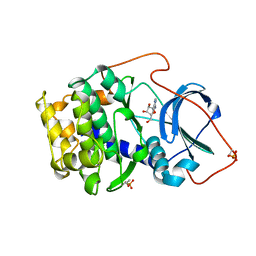 | | A BINARY COMPLEX OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND ADENOSINE FURTHER DEFINES CONFORMATIONAL FLEXIBILITY | | Descriptor: | ADENOSINE MONOPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Cox, S, Xuong, N, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A binary complex of the catalytic subunit of cAMP-dependent protein kinase and adenosine further defines conformational flexibility.
Structure, 5, 1997
|
|
1BFE
 
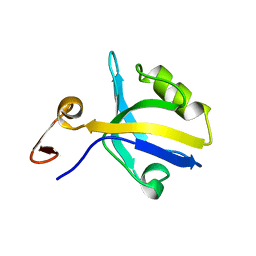 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 | | Descriptor: | PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
7G84
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1102357527 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, N-[(3R)-6-oxopiperidin-3-yl]-1,3-thiazole-4-carboxamide, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G8E
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z104479710 | | Descriptor: | 1-(1H-benzimidazol-2-yl)methanamine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
