2JAD
 
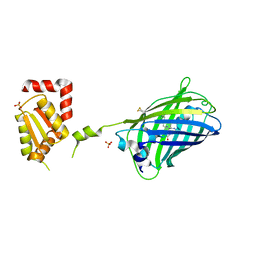 | |
6YOV
 
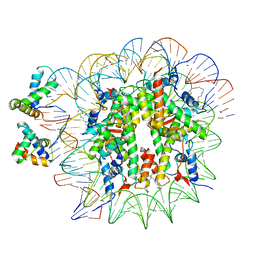 | | OCT4-SOX2-bound nucleosome - SHL+6 | | Descriptor: | DNA (142-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6WVI
 
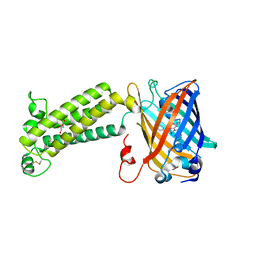 | | VKOR-like from Takifugu rubripes | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Vitamin K epoxide reductase-like protein, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
8DFL
 
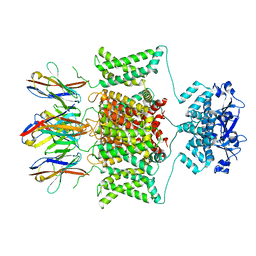 | |
2P4M
 
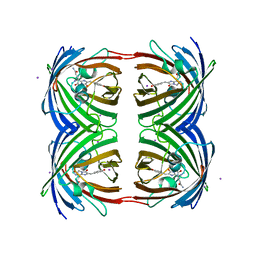 | | High pH structure of Rtms5 H146S variant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Wilmann, P.G, Olsen, S, Byres, E, Smith, S.C, Dove, S.G, Turcic, K.N, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for the pH-dependent increase in fluorescence efficiency of chromoproteins
J.Mol.Biol., 368, 2007
|
|
8F6P
 
 | | Rat Cardiac Sodium Channel with Ranolazine Bound | | Descriptor: | (R)-ranolazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lenaeus, M.J, Tonggu, L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the cardiac sodium channel by the atypical antiarrhythmic drug ranolazine.
Nat Cardiovasc Res, 2, 2023
|
|
8XTX
 
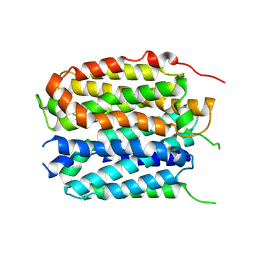 | | structure of a protein | | Descriptor: | Green fluorescent protein,Vesicular acetylcholine transporter,antibody | | Authors: | Zhao, Y, Ma, Q, Dong, Y, Meng, Y. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-25 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Binding mechanism and antagonism of the vesicular acetylcholine transporter VAChT.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8XTW
 
 | | structure of a protein | | Descriptor: | ACETYLCHOLINE, Green fluorescent protein,Vesicular acetylcholine transporter,antibody | | Authors: | Zhao, Y, Ma, Q, Dong, Y, Meng, Y. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-25 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Binding mechanism and antagonism of the vesicular acetylcholine transporter VAChT.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8XTY
 
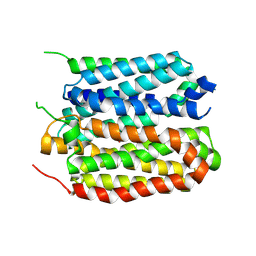 | | structure of a protein | | Descriptor: | Green fluorescent protein,Vesicular acetylcholine transporter,Green fluorescent protein,Vesicular acetylcholine transporter,Green fluorescent protein,Vesicular acetylcholine transporter,Green fluorescent protein,Vesicular acetylcholine transporter,antibody, vesamicol | | Authors: | Zhao, Y, Ma, Q, Dong, Y, Meng, Y. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-25 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Binding mechanism and antagonism of the vesicular acetylcholine transporter VAChT.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6GEL
 
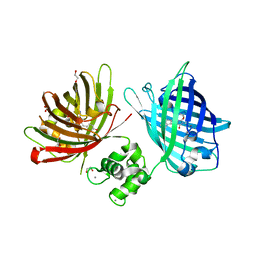 | | The structure of TWITCH-2B | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-26 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
6GEZ
 
 | | THE STRUCTURE OF TWITCH-2B N532F | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
9J97
 
 | | Closed structure of human XPR1 | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, CHOLESTEROL, PHOSPHATE ION, ... | | Authors: | Wang, Y, Wang, Y, Yang, H, Shen, H. | | Deposit date: | 2024-08-22 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of phosphate export by human XPR1.
Cell Res., 35, 2025
|
|
9J98
 
 | | Open structure of human XPR1 | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, CHOLESTEROL, Solute carrier family 53 member 1,Green fluorescent protein | | Authors: | Wang, Y, Wang, Y, Yang, H, Shen, H. | | Deposit date: | 2024-08-22 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural basis of phosphate export by human XPR1.
Cell Res., 35, 2025
|
|
1MOV
 
 | | Crystal structure of Coral protein mutant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | Deposit date: | 2002-09-10 | | Release date: | 2003-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
1KYS
 
 | | Crystal Structure of a Zn-bound Green Fluorescent Protein Biosensor | | Descriptor: | Green Fluorescent Protein, ZINC ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-02-05 | | Release date: | 2002-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural chemistry of a green fluorescent protein Zn biosensor.
J.Am.Chem.Soc., 124, 2002
|
|
1KYP
 
 | | Crystal Structure of an Apo Green Fluorescent Protein Zn Biosensor | | Descriptor: | Green Fluorescent Protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-02-05 | | Release date: | 2002-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural chemistry of a green fluorescent protein Zn biosensor.
J.Am.Chem.Soc., 124, 2002
|
|
3UFZ
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the universal genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
1MOU
 
 | | Crystal structure of Coral pigment | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | Deposit date: | 2002-09-10 | | Release date: | 2003-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
3UG0
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the simplified genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
8J0J
 
 | | AtSLAC1 8D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
6QQ8
 
 | |
6QQA
 
 | |
6QQB
 
 | |
6QQ9
 
 | |
8J1E
 
 | | AtSLAC1 in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
