7XAE
 
 | | Crystal strucutre of PD-L1 and 3ONJA protein | | Descriptor: | 2IC6, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Crystal strucutre of PD-L1 and 2IC6 protein
To Be Published
|
|
1DR9
 
 | | CRYSTAL STRUCTURE OF A SOLUBLE FORM OF B7-1 (CD80) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T LYMPHOCYTE ACTIVATION ANTIGEN | | Authors: | Ikemizu, S, Jones, E.Y, Stuart, D.I, Davis, S.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and dimerization of a soluble form of B7-1.
Immunity, 12, 2000
|
|
1E07
 
 | |
1F97
 
 | | SOLUBLE PART OF THE JUNCTION ADHESION MOLECULE FROM MOUSE | | Descriptor: | JUNCTION ADHESION MOLECULE, MAGNESIUM ION | | Authors: | Kostrewa, D, Brockhaus, M, D'Arcy, A, Dale, G, Bazzoni, G, Dejana, E, Winkler, F, Hennig, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif.
EMBO J., 20, 2001
|
|
1I8L
 
 | | HUMAN B7-1/CTLA-4 CO-STIMULATORY COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOTOXIC T-LYMPHOCYTE PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD80, ... | | Authors: | Stamper, C.C, Somers, W.S, Mosyak, L. | | Deposit date: | 2001-03-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses.
Nature, 410, 2001
|
|
3AB0
 
 | |
3ALP
 
 | | Cell adhesion protein | | Descriptor: | CITRIC ACID, HEXANE-1,6-DIOL, Poliovirus receptor-related protein 1 | | Authors: | Narita, H, Nakagawa, A, Suzuki, M. | | Deposit date: | 2010-08-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal Structure of the cis-Dimer of Nectin-1: implications for the architecture of cell-cell junctions
J.Biol.Chem., 286, 2011
|
|
3BIS
 
 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BP5
 
 | | Crystal structure of the mouse PD-1 and PD-L2 complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Yan, Q, Lazar-Molnar, E, Cao, E, Ramagopal, U.A, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-18 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BP6
 
 | | Crystal structure of the mouse PD-1 Mutant and PD-L2 complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Yan, Q, Lazar-Molnar, E, Cao, E, Ramagopal, U.A, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-18 | | Release date: | 2009-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the mouse PD-1 A99L and PD-L2 complex
To be published
|
|
6X4T
 
 | | Crystal structure of ICOS-L in complex with Prezalumab and VNAR domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Rujas, E, Sicard, T, Julien, J.P. | | Deposit date: | 2020-05-23 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of the ICOS/ICOS-L immune complex reveals high molecular mimicry by therapeutic antibodies.
Nat Commun, 11, 2020
|
|
3BIK
 
 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6DLE
 
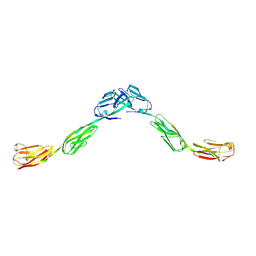 | | Crystal structure of IgLON5 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgLON family member 5 | | Authors: | Ranaivoson, F.M, Turk, L.S, Ozkan, E, Montelione, G.T, Comoletti, D. | | Deposit date: | 2018-06-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.994 Å) | | Cite: | Structure of a heterodimer of neuronal cell surface proteins
Structure, 2019
|
|
6C6Q
 
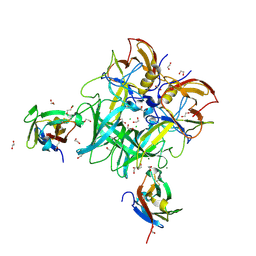 | |
6E47
 
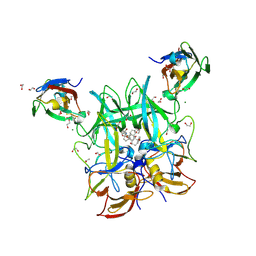 | |
6E48
 
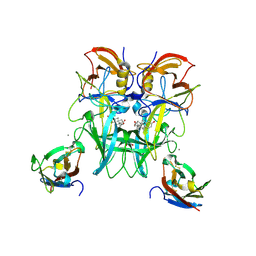 | | Crystal Structure of the Murine Norovirus VP1 P domain in complex with the CD300lf Receptor and Lithocholic Acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EG1
 
 | | Crystal structure of Dpr2 Ig1-Ig2 in complex with DIP-Theta Ig1-Ig3 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cosmanescu, F, Patel, S, Shapiro, L. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
6AVZ
 
 | | Crystal structure of the HopQ-CEACAM3 WT complex | | Descriptor: | CALCIUM ION, Carcinoembryonic antigen-related cell adhesion molecule 3, HopQ, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6AW3
 
 | | Crystal structure of the HopQ-CEACAM3 L44Q complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 3, HopQ | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
2PTT
 
 | |
6ARQ
 
 | | Crystal structure of CD96 (D1) bound to CD155/necl-5 (D1-3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, T-cell surface protein tactile, ... | | Authors: | Deuss, F.A, Watson, G.M, Rossjohn, J, Berry, R. | | Deposit date: | 2017-08-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis for CD96 Immune Receptor Recognition of Nectin-like Protein-5, CD155.
Structure, 27, 2019
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6AW2
 
 | | Crystal structure of the HopQ-CEACAM1 complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, HopQ | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
2OZ4
 
 | | Structural Plasticity in IgSF Domain 4 of ICAM-1 Mediates Cell Surface Dimerization | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB FRAGMENT LIGHT CHAIN, ... | | Authors: | Chen, X, Kim, T.D, Carman, C.V, Mi, L, Song, G, Springer, T.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity in Ig superfamily domain 4 of ICAM-1 mediates cell surface dimerization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
