7ZUW
 
 | | Structure of RQT (C1) bound to the stalled ribosome in a disome unit from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZNJ
 
 | | Structure of an ALYREF-exon junction complex hexamer | | Descriptor: | Eukaryotic initiation factor 4A-III, N-terminally processed, MAGNESIUM ION, ... | | Authors: | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | Deposit date: | 2022-04-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
7ZNK
 
 | | Structure of an endogenous human TREX complex bound to mRNA | | Descriptor: | RNA, Spliceosome RNA helicase DDX39B, THO complex subunit 1, ... | | Authors: | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | Descriptor: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
5LQW
 
 | | yeast activated spliceosome | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-processing protein 45, Pre-mRNA-splicing factor 8, ... | | Authors: | Rauhut, R, Luehrmann, R. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Molecular architecture of the Saccharomyces cerevisiae activated spliceosome
Science, 6306, 2016
|
|
5MC6
 
 | | Cryo-EM structure of a native ribosome-Ski2-Ski3-Ski8 complex from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Schmidt, C, Kowalinski, E, Shanmuganathan, V, Defenouillere, Q, Braunger, K, Heuer, A, Pech, M, Namane, A, Berninghausen, O, Fromont-Racine, M, Jacquier, A, Conti, E, Becker, T, Beckmann, R. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-18 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex.
Science, 354, 2016
|
|
5MQF
 
 | | Cryo-EM structure of a human spliceosome activated for step 2 of splicing (C* complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ATP-dependent RNA helicase DHX8, Cell division cycle 5-like protein, ... | | Authors: | Bertram, K, Hartmuth, K, Kastner, B. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structure of a human spliceosome activated for step 2 of splicing.
Nature, 542, 2017
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
7ZPQ
 
 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C1) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZRS
 
 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C2) - composite map | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
8B9L
 
 | | Cryo-EM structure of MLE | | Descriptor: | Dosage compensation regulator | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
8B9K
 
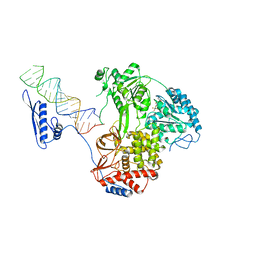 | |
8B9J
 
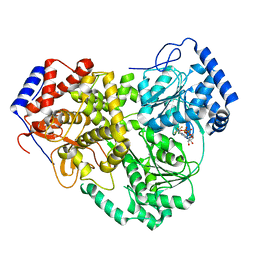 | | Cryo-EM structure of MLE in complex with ADP:AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dosage compensation regulator, MAGNESIUM ION, ... | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
8B9I
 
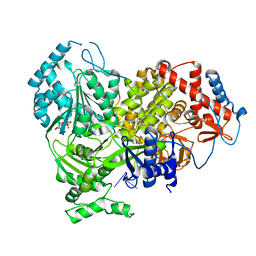 | |
8B9G
 
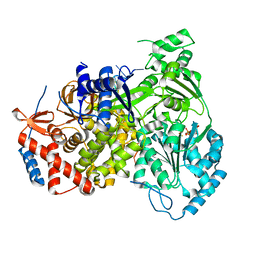 | | Cryo-EM structure of MLE in complex with ADP:AlF4 and U10 RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dosage compensation regulator, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
8CH6
 
 | | Structure of a late-stage activated spliceosome (BAqr) arrested with a dominant-negative Aquarius mutant (state B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Cretu, C, Schmitzova, J, Pena, V. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of catalytic activation in human splicing.
Nature, 617, 2023
|
|
7AJU
 
 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
7AJT
 
 | | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
6ZQE
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-A (Poly-Ala) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
7A5P
 
 | | Human C Complex Spliceosome - Medium-resolution PERIPHERY | | Descriptor: | Cell division cycle 5-like protein, Crooked neck-like protein 1, Eukaryotic initiation factor 4A-III, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
6ZQG
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-C | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
7AOI
 
 | | Trypanosoma brucei mitochondrial ribosome large subunit assembly intermediate | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Tobiasson, V, Gahura, O, Aibara, S, Baradaran, R, Zikova, A, Amunts, A. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-02 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Interconnected assembly factors regulate the biogenesis of mitoribosomal large subunit.
Embo J., 40, 2021
|
|
6ZMW
 
 | | Structure of a human 48S translational initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6ZQD
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Post-A1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZYM
 
 | | Human C Complex Spliceosome - High-resolution CORE | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Corepressor interacting with RBPJ 1, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
