7CMF
 
 | | Crystal structure of human P-cadherin REC12 (monomer) in complex with 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine (inhibitor) | | Descriptor: | 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3 | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of cadherin dimerization by chemical fragments as a trigger to inhibit cell adhesion
Commun Biol, 4, 2021
|
|
8FPX
 
 | |
7CKN
 
 | |
4NIF
 
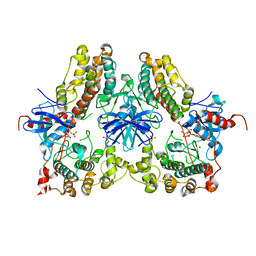 | | Heterodimeric structure of ERK2 and RSK1 | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-1, ... | | Authors: | Gogl, G, Remenyi, A. | | Deposit date: | 2013-11-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural assembly of the signaling competent ERK2-RSK1 heterodimeric protein kinase complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8FPW
 
 | |
7MRO
 
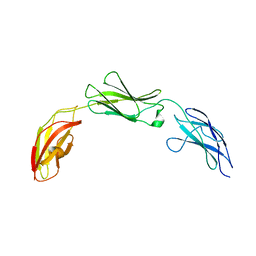 | | Zebrafish CNTN4 FN1-FN3 domains | | Descriptor: | Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7UF9
 
 | | CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-{[(2R)-1-oxo-3-phenyl-1-{[2-(pyridin-4-yl)ethyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
6E5E
 
 | |
7COO
 
 | |
7MFZ
 
 | |
7CON
 
 | |
7UFF
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, N-(2-oxo-2-{[(2S)-1-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]amino}ethyl)pyridine-3-carboxamide, PHOSPHATE ION, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
4NJT
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
7UFD
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | (2S)-3-phenyl-2-({(2S)-3-phenyl-2-[3-(pyridin-3-yl)propanamido]propyl}sulfanyl)-N-[2-(pyridin-3-yl)ethyl]propanamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
7CP8
 
 | |
7UFE
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, GLYCEROL, N-[(2S)-1-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]pyridine-3-carboxamide, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
6E5Q
 
 | |
6E5T
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiorcin (Abn-CBDO) | | Descriptor: | (1'R,2'R)-5',6-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
4NKV
 
 | |
5W5Q
 
 | | MAP4K4 in complex with inhibitor compound 12 (N3-methyl-10-(3-methyl-3-(5-methyloxazol-2-yl)but-1-yn-1-yl)-6,7-dihydro-5H-5,7-methanobenzo[c]imidazo[1,2-a]azepine-2,3-dicarboxamide) | | Descriptor: | (5s,7s)-N~3~-methyl-10-[3-methyl-3-(5-methyl-1,3-oxazol-2-yl)but-1-yn-1-yl]-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2017-06-15 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure Based Design of Potent Selective Inhibitors of Protein Kinase D1 (PKD1).
Acs Med.Chem.Lett., 10, 2019
|
|
6A17
 
 | | Crystal structure of CYP90B1 in complex with brassinazole | | Descriptor: | (2R,3S)-4-(4-chlorophenyl)-2-phenyl-3-(1H-1,2,4-triazol-1-yl)butan-2-ol, CHLORIDE ION, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
7UFA
 
 | | CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-4-yl)propyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
7MFX
 
 | |
4NM0
 
 | | Crystal structure of peptide inhibitor-free GSK-3/Axin complex | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE-5'-DIPHOSPHATE, Axin-1, ... | | Authors: | Chu, M.L.-H, Stamos, J.L, Enos, M.D, Shah, N, Weis, W.I. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6.
Elife, 3, 2014
|
|
7MFY
 
 | |
