1SG4
 
 | | Crystal structure of human mitochondrial delta3-delta2-enoyl-CoA isomerase | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, mitochondrial, OCTANOYL-COENZYME A | | Authors: | Partanen, S.T, Novikov, D.K, Popov, A.N, Mursula, A.M, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2004-02-23 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of human mitochondrial Delta3-Delta2-enoyl-CoA isomerase shows a novel mode of binding for the fatty acyl group.
J.Mol.Biol., 342, 2004
|
|
1SO0
 
 | | Crystal structure of human galactose mutarotase complexed with galactose | | Descriptor: | aldose 1-epimerase, beta-D-galactopyranose | | Authors: | Thoden, J.B, Timson, D.J, Reece, R.J, Holden, H.M. | | Deposit date: | 2004-03-12 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular structure of human galactose mutarotase
J.Biol.Chem., 279, 2004
|
|
1DEA
 
 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1S2W
 
 | | Crystal structure of phosphoenolpyruvate mutase in high ionic strength | | Descriptor: | Phosphoenolpyruvate phosphomutase, SULFATE ION | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1EQJ
 
 | | CRYSTAL STRUCTURE OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, PHOSPHOGLYCERATE MUTASE | | Authors: | Jedrzejas, M.J, Chander, M, Setlow, P, Krishnasamy, G. | | Deposit date: | 2000-04-05 | | Release date: | 2001-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of catalysis of the cofactor-independent phosphoglycerate mutase from Bacillus stearothermophilus. Crystal structure of the complex with 2-phosphoglycerate.
J.Biol.Chem., 275, 2000
|
|
1SQC
 
 | |
1S2U
 
 | | Crystal structure of the D58A phosphoenolpyruvate mutase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1E59
 
 | |
1FKK
 
 | | ATOMIC STRUCTURE OF FKBP12, AN IMMUNOPHILIN BINDING PROTEIN | | Descriptor: | FK506 BINDING PROTEIN, SULFATE ION | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1FAP
 
 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | FK506-BINDING PROTEIN, FRAP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Choi, J, Chen, J, Schreiber, S.L, Clardy, J. | | Deposit date: | 1996-03-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP.
Science, 273, 1996
|
|
1TRE
 
 | |
1EJJ
 
 | | CRYSTAL STRUCTURAL ANALYSIS OF PHOSPHOGLYCERATE MUTASE COCRYSTALLIZED WITH 3-PHOSPHOGLYCERATE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, PHOSPHOGLYCERATE MUTASE | | Authors: | Jedrzejas, M.J, Chander, M, Setlow, P, Krishnasamy, G. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of action of a novel phosphoglycerate mutase from Bacillus stearothermophilus.
EMBO J., 19, 2000
|
|
1BJX
 
 | | HUMAN PROTEIN DISULFIDE ISOMERASE, NMR, 24 STRUCTURES | | Descriptor: | PROTEIN DISULFIDE ISOMERASE | | Authors: | Kemmink, J, Dijkstra, K, Mariani, M, Scheek, R.M, Penka, E, Nilges, M, Darby, N.J. | | Deposit date: | 1998-06-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure in solution of the b domain of protein disulfide isomerase.
J.Biomol.NMR, 13, 1999
|
|
1PMI
 
 | | Candida Albicans Phosphomannose Isomerase | | Descriptor: | PHOSPHOMANNOSE ISOMERASE, ZINC ION | | Authors: | Cleasby, A, Skarzynski, T, Wonacott, A, Davies, G.J, Hubbard, R.E, Proudfoot, A.E.I, Wells, T.N.C, Payton, M.A, Bernard, A.R. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The x-ray crystal structure of phosphomannose isomerase from Candida albicans at 1.7 angstrom resolution.
Nat.Struct.Biol., 3, 1996
|
|
1TPE
 
 | |
1TTJ
 
 | |
1FKL
 
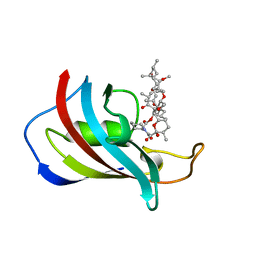 | | ATOMIC STRUCTURE OF FKBP12-RAPAYMYCIN, AN IMMUNOPHILIN-IMMUNOSUPPRESSANT COMPLEX | | Descriptor: | FK506 BINDING PROTEIN, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1FKJ
 
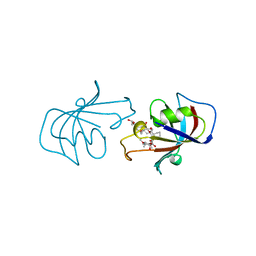 | | ATOMIC STRUCTURE OF FKBP12-FK506, AN IMMUNOPHILIN IMMUNOSUPPRESSANT COMPLEX | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1TM0
 
 | | Crystal Structure of the putative proline racemase from Brucella melitensis, Northeast Structural Genomics Target LR31 | | Descriptor: | PROLINE RACEMASE | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Ho, C.K, Ma, L.-C, Cooper, B, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-10 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1TPF
 
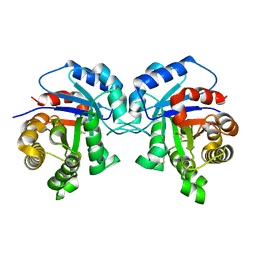 | |
1FZT
 
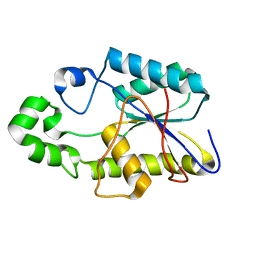 | | SOLUTION STRUCTURE AND DYNAMICS OF AN OPEN B-SHEET, GLYCOLYTIC ENZYME-MONOMERIC 23.7 KDA PHOSPHOGLYCERATE MUTASE FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | PHOSPHOGLYCERATE MUTASE | | Authors: | Uhrinova, S, Uhrin, D, Nairn, J, Price, N.C, Fothergill-Gilmore, L.A. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of an open beta-sheet, glycolytic enzyme, monomeric 23.7 kDa phosphoglycerate mutase from Schizosaccharomyces pombe.
J.Mol.Biol., 306, 2001
|
|
1TTI
 
 | |
1TRI
 
 | |
2DJK
 
 | |
2DWU
 
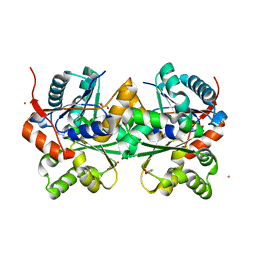 | | Crystal Structure of Glutamate Racemase Isoform RacE1 from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, GLYCEROL, Glutamate racemase, ... | | Authors: | Mehboob, S, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2006-08-17 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Analysis of Two Glutamate Racemase Isozymes from Bacillus anthracis and Implications for Inhibitor Design
J.Mol.Biol., 371, 2007
|
|
