8KE8
 
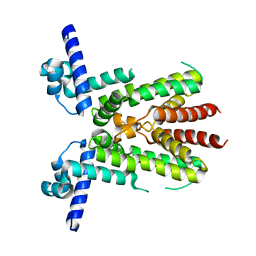 | | Crystal structure of TetR-type transcriptional factor NalC from P. aeruginosa | | Descriptor: | NalC | | Authors: | Lee, J.Y, Jeong, K.H, Ko, J.H, Son, S.B. | | Deposit date: | 2023-08-11 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the transcriptional regulator NalC, a key component of the MexAB-OprM efflux pump system, from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 679, 2023
|
|
8I4B
 
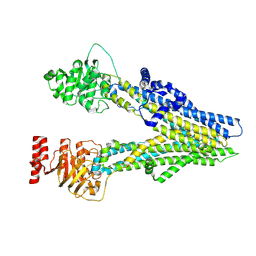 | | Cryo-EM structure of apo-form ABCC4 | | Descriptor: | ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4A
 
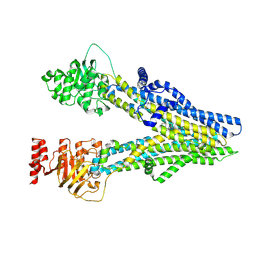 | | Cryo-EM structure of dipyridamole-bound ABCC4 | | Descriptor: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4C
 
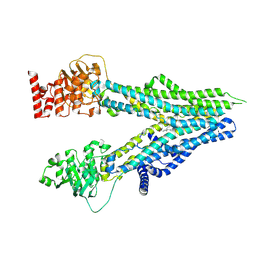 | | Cryo-EM structure of U46619-bound ABCC4 | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8J3Z
 
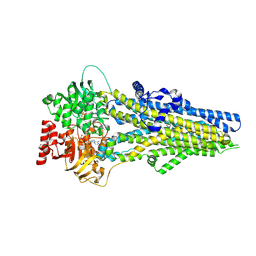 | | Cryo-EM structure of ATP-U46619-bound ABCC4 | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, ... | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-04-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8J3W
 
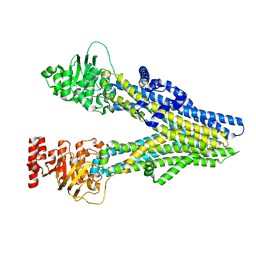 | | Cryo-EM structure of aspirin-bound ABCC4 | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-04-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
6NNG
 
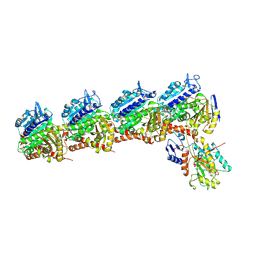 | | Tubulin-RB3_SLD-TTL in complex with compound DJ95 | | Descriptor: | 2-(1H-indol-6-yl)-4-(3,4,5-trimethoxyphenyl)-1H-imidazo[4,5-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-01-15 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Colchicine Binding Site Agent DJ95 Overcomes Drug Resistance and Exhibits Antitumor Efficacy.
Mol.Pharmacol., 96, 2019
|
|
7NIU
 
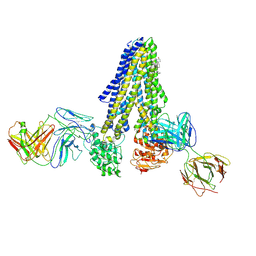 | |
7NIV
 
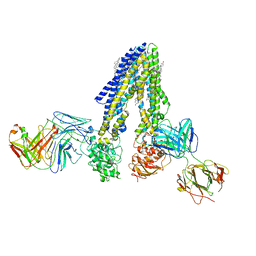 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (phosphatidylcholine-bound, occluded conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment light chain, CHOLESTEROL, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NIW
 
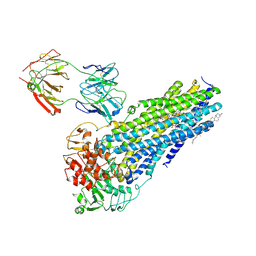 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab (posaconazole-bound, inward-open conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment heavy chain, 4B1 Fab-fragment light chain, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2AP2
 
 | |
5T83
 
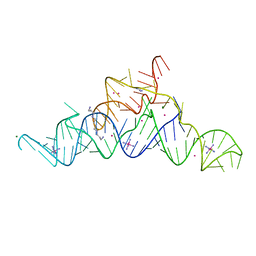 | | Structure of a guanidine-I riboswitch from S. acidophilus | | Descriptor: | GUANIDINE, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Reiss, C.W, Xiong, Y, Strobel, S.A. | | Deposit date: | 2016-09-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Basis for Ligand Binding to the Guanidine-I Riboswitch.
Structure, 25, 2017
|
|
7A09
 
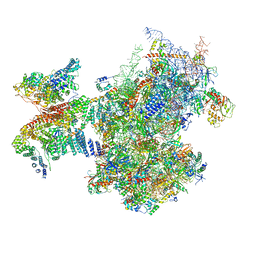 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State III | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-08-07 | | Release date: | 2020-10-14 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
6ZME
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-eERF1 ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZVJ
 
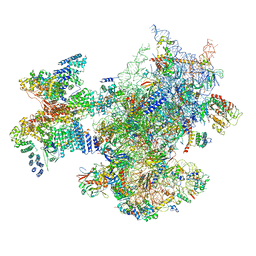 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State II | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
7JU3
 
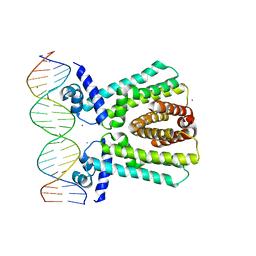 | | MtrR bound to the mtrCDE operator from Neisseria gonorrhoeae | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*AP*TP*AP*CP*AP*CP*GP*AP*TP*TP*GP*CP*AP*CP*GP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*CP*CP*GP*TP*GP*CP*AP*AP*TP*CP*GP*TP*GP*TP*AP*TP*GP*T)-3'), ... | | Authors: | Beggs, G.A, Shafer, W.M, Brennan, R.G. | | Deposit date: | 2020-08-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Neisseria gonorrhoeae MtrR-operator complexes reveal molecular mechanisms of DNA recognition and antibiotic resistance-conferring clinical mutations.
Nucleic Acids Res., 49, 2021
|
|
7JNP
 
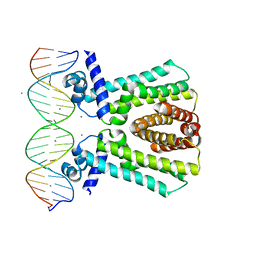 | | MtrR bound to the rpoH operator from Neisseria gonorrhoeae | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*AP*CP*AP*TP*AP*CP*GP*TP*GP*GP*TP*TP*GP*TP*AP*TP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*AP*TP*AP*CP*AP*AP*CP*CP*AP*CP*GP*TP*AP*TP*GP*TP*A)-3'), ... | | Authors: | Beggs, G.A, Shafer, W.M, Brennan, R.G. | | Deposit date: | 2020-08-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of Neisseria gonorrhoeae MtrR-operator complexes reveal molecular mechanisms of DNA recognition and antibiotic resistance-conferring clinical mutations.
Nucleic Acids Res., 49, 2021
|
|
4ESF
 
 | |
4ESB
 
 | | Crystal structure of PadR-like transcriptional regulator (BC4206) from Bacillus cereus strain ATCC 14579 | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Fibriansah, G, Kovacs, A.T, Kuipers, O.P, Thunnissen, A.M.W.H. | | Deposit date: | 2012-04-23 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Two Transcriptional Regulators from Bacillus cereus Define the Conserved Structural Features of a PadR Subfamily.
Plos One, 7, 2012
|
|
1BLN
 
 | | ANTI-P-GLYCOPROTEIN FAB MRK-16 | | Descriptor: | PROTEIN (MONOCLONAL ANTIBODY MRK-16 (HEAVY CHAIN)), PROTEIN (MONOCLONAL ANTIBODY MRK-16 (LIGHT CHAIN)) | | Authors: | Vasudevan, S, Tsuruo, T, Rose, D.R. | | Deposit date: | 1998-07-16 | | Release date: | 1998-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mode of binding of anti-P-glycoprotein antibody MRK-16 to its antigen. A crystallographic and molecular modeling study.
J.Biol.Chem., 273, 1998
|
|
6MH4
 
 | |
6MH5
 
 | |
4U8D
 
 | |
6WXI
 
 | | Colicin E1 fragment in nanodisc-embedded TolC | | Descriptor: | Outer membrane protein TolC | | Authors: | Kaelber, J.T, Budiardjo, S.J, Firlar, E, Ikujuni, A.P, Slusky, J.S.G. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Colicin E1 opens its hinge to plug TolC.
Elife, 11, 2022
|
|
6WXH
 
 | | Colicin E1 fragment in nanodisc-embedded TolC | | Descriptor: | Colicin-E1, Outer membrane protein TolC | | Authors: | Kaelber, J.T, Budiardjo, S.J, Firlar, E, Case, D.A, Ikujuni, A.P, Slusky, J.S.G. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Colicin E1 opens its hinge to plug TolC.
Elife, 11, 2022
|
|
