2VHK
 
 | |
6Y1P
 
 | | Human Aldose Reductase Mutant L300/301A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid) | | Descriptor: | 3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid, Aldo-keto reductase family 1 member B1, CITRIC ACID, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2020-02-13 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Human Aldose Reductase Mutant L300/301A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid)
To Be Published
|
|
2ZQ7
 
 | | Apo structure of Class A beta-lactamase Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | Beta-lactamase Toho-1, SULFATE ION | | Authors: | Shimamura, T, Nitanai, Y, Uchiyama, T, Ago, H, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Improvement of the Crystal Quality by the Surface Mutations on a Beta-Lactamase Toho-1
To be Published
|
|
3QR7
 
 | | Crystal structure of the C-terminal fragment of the bacteriophage P2 membrane-piercing protein gpV | | Descriptor: | Baseplate assembly protein V, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Browning, C, Shneider, M, Leiman, P.G. | | Deposit date: | 2011-02-17 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Phage pierces the host cell membrane with the iron-loaded spike.
Structure, 20, 2012
|
|
1A7Y
 
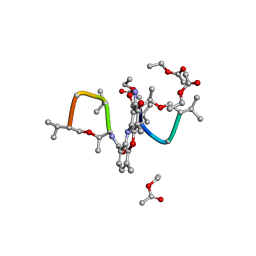 | | CRYSTAL STRUCTURE OF ACTINOMYCIN D | | Descriptor: | ACTINOMYCIN D, ETHYL ACETATE, METHANOL | | Authors: | Schafer, M, Sheldrick, G.M, Bahner, I, Lackner, H. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
1NLS
 
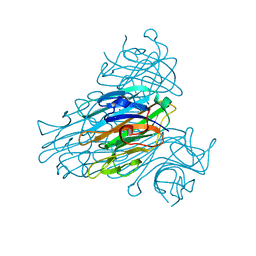 | | CONCANAVALIN A AND ITS BOUND SOLVENT AT 0.94A RESOLUTION | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Deacon, A.M, Gleichmann, T, Helliwell, J.R, Kalb(Gilboa), A.J. | | Deposit date: | 1997-01-28 | | Release date: | 1997-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Structure of Concanavalin a and its Bound Solvent Determined with Small-Molecule Accuracy at 0.94 A Resolution
J.Chem.Soc.,Faraday Trans., 93, 1997
|
|
1ZFF
 
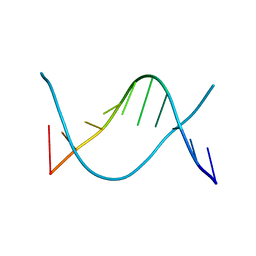 | | TTC Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*TP*TP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2V8B
 
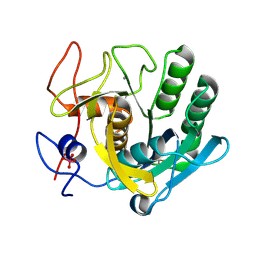 | |
4BY8
 
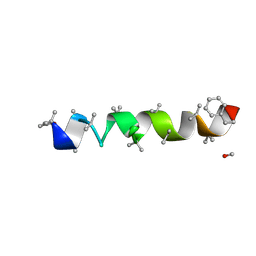 | |
1GVU
 
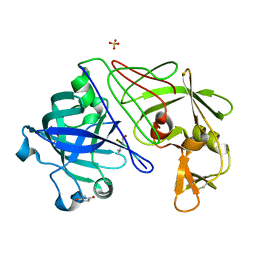 | | Endothiapepsin complex with H189 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR, H189, ... | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
2PPP
 
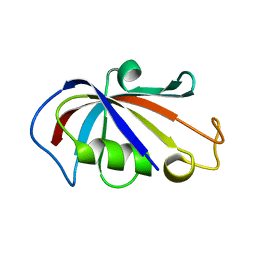 | |
2GLK
 
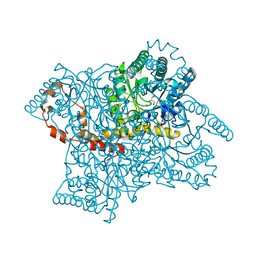 | | High-resolution study of D-Xylose isomerase, 0.94A resolution. | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Carrell, H.L, Hanson, B.L, Harp, J.M, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5RVI
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000084843283 | | Descriptor: | CHLORZOXAZONE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2GUD
 
 | |
1GVK
 
 | | Porcine pancreatic elastase acyl enzyme at 0.95 A resolution | | Descriptor: | CALCIUM ION, ELASTASE 1, PEPTIDE INHIBITOR, ... | | Authors: | Katona, G, Wilmouth, R.C, Wright, P.A, Berglund, G.I, Hajdu, J, Neutze, R, Schofield, C.J. | | Deposit date: | 2002-02-14 | | Release date: | 2002-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | X-Ray Structure of a Serine Protease Acyl-Enzyme Complex at 0.95-A Resolution.
J.Biol.Chem., 277, 2002
|
|
2FDN
 
 | | 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDI-URICI | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Meyer, J, Moulis, J.M. | | Deposit date: | 1997-10-01 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Atomic resolution (0.94 A) structure of Clostridium acidurici ferredoxin. Detailed geometry of [4Fe-4S] clusters in a protein.
Biochemistry, 36, 1997
|
|
6RZH
 
 | | Galectin-3C in complex with para-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(4-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.947 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
6RH4
 
 | | Human Carbonic Anhydrase II in complex with 4-Nitrobenzenesulfonamide. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-nitrobenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.948 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
4MZC
 
 | | Atomic Resolution Structure of PfGrx1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Glutaredoxin | | Authors: | Yogavel, M, Sharma, A. | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.949 Å) | | Cite: | Atomic resolution crystal structure of glutaredoxin 1 from Plasmodium falciparum and comparison with other glutaredoxins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5R3C
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 36, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.949 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
2YL0
 
 | |
7FXF
 
 | | Crystal Structure of human FABP4 in complex with 2-[(2-chlorophenoxy)methyl]-1,3-thiazole-4-carboxylic acid | | Descriptor: | 2-[(2-chlorophenoxy)methyl]-1,3-thiazole-4-carboxylic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
4G9S
 
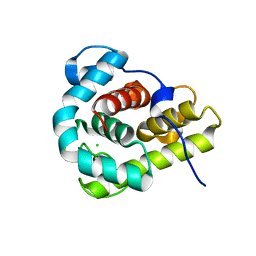 | | Crystal structure of Escherichia coli PliG in complex with Atlantic salmon g-type lysozyme | | Descriptor: | CHLORIDE ION, CITRATE ANION, Goose-type lysozyme, ... | | Authors: | Leysen, S, Vanderkelen, L, Weeks, S.D, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-07-24 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis of bacterial defense against g-type lysozyme-based innate immunity.
Cell.Mol.Life Sci., 70, 2013
|
|
7FWK
 
 | | Crystal Structure of human FABP4 in complex with 1-[(2-chlorophenyl)methyl]pyrrole-2-carboxamide | | Descriptor: | 1-[(2-chlorophenyl)methyl]pyrrole-2-carboxamide, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
1N1P
 
 | |
