8GFQ
 
 | |
8GFD
 
 | |
8GEZ
 
 | |
8GF0
 
 | |
8GFP
 
 | | Crystal structure of soluble lytic transglycosylase Cj0843 of Campylobacter jejuni in complex with N-acetyl-2,3-dehydro-2-deoxyneuraminic acid inhibitor | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CITRIC ACID, Lytic transglycosylase domain-containing protein, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Exploring the inhibition of the soluble lytic transglycosylase Cj0843c of Campylobacter jejuni via targeting different sites with different scaffolds.
Protein Sci., 32, 2023
|
|
8GFI
 
 | |
8GFS
 
 | | Crystal structure of soluble lytic transglycosylase Cj0843 of Campylobacter jejuni in complex with siastatin B inhibitor | | Descriptor: | (2S,3R,4S,5S)-2-(acetylamino)-5-carboxy-3,4-dihydroxypiperidinium, CITRIC ACID, Lytic transglycosylase domain-containing protein | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Exploring the inhibition of the soluble lytic transglycosylase Cj0843c of Campylobacter jejuni via targeting different sites with different scaffolds.
Protein Sci., 32, 2023
|
|
8GFB
 
 | |
8GFL
 
 | |
8GR2
 
 | |
5TPV
 
 | | X-ray structure of WlaRA (TDP-fucose-3,4-ketoisomerase) from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, THYMIDINE-5'-DIPHOSPHATE, WlaRA, ... | | Authors: | Holden, H.M, Thoden, J.B, Li, Z.A, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinograd, E, Schoenhofen, I.C, Gilbert, M, Li, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
5TPU
 
 | | x-ray structure of the WlaRB TDP-quinovose 3,4-ketoisomerase from campylobacter jejuni | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Holden, H.M, Thoden, J.B, Li, J.Z, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinogradov, E, Schoenhofen, I.C, Gilbert, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
3QQT
 
 | |
3ORD
 
 | |
4W92
 
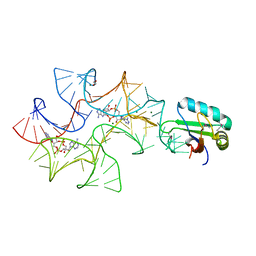 | | Crystal structure of Bacillus subtilis cyclic-di-AMP riboswitch ydaO | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, 1,2-ETHANEDIOL, C-di-AMP ribsoswitch, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA.
Embo J., 33, 2014
|
|
9NTE
 
 | | Helix pomatia AMP deaminase (HPAMPD) apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
9NTF
 
 | | Helix pomatia AMP deaminase (HPAMPD) with unknown density in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
9NTH
 
 | | Helix pomatia AMP deaminase (HPAMPD) in complex with Pentostatin monophosphate (PMP) | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
5JRH
 
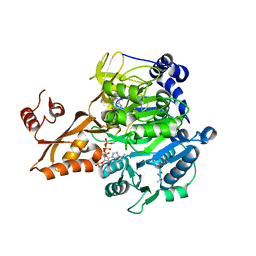 | | Crystal structure of Salmonella enterica acetyl-CoA synthetase (Acs) in complex with cAMP and Coenzyme A | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Acetyl-coenzyme A synthetase, ... | | Authors: | Shen, L, Zhang, Y. | | Deposit date: | 2016-05-06 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Cyclic AMP Inhibits the Activity and Promotes the Acetylation of Acetyl-CoA Synthetase through Competitive Binding to the ATP/AMP Pocket.
J. Biol. Chem., 292, 2017
|
|
4CYD
 
 | | GlxR bound to cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, PROBABLE EXPRESSION TAG, ... | | Authors: | Townsend, P.D, Bott, M, Cann, M.J, Pohl, E. | | Deposit date: | 2014-04-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structures of Apo and Camp-Bound Glxr from Corynebacterium Glutamicum Reveal Structural and Dynamic Changes Upon Camp Binding in Crp/Fnr Family Transcription Factors.
Plos One, 9, 2014
|
|
3H3U
 
 | | Crystal structure of CRP (cAMP receptor Protein) from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY CRP/FNR-FAMILY), SULFATE ION | | Authors: | Kumar, P, Joshi, D.C, Akif, M, Akhter, Y, Hasnain, S.E, Mande, S.C. | | Deposit date: | 2009-04-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mapping conformational transitions in cyclic AMP receptor protein: crystal structure and normal-mode analysis of Mycobacterium tuberculosis apo-cAMP receptor protein
Biophys.J., 98, 2010
|
|
4QK9
 
 | | Thermovirga lienii c-di-AMP riboswitch | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, C-di-AMP riboswitch, MAGNESIUM ION | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2014-06-05 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into recognition of c-di-AMP by the ydaO riboswitch.
Nat.Chem.Biol., 10, 2014
|
|
4QK8
 
 | | Thermoanaerobacter pseudethanolicus c-di-AMP riboswitch | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, C-di-AMP riboswitch, MAGNESIUM ION, ... | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2014-06-05 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into recognition of c-di-AMP by the ydaO riboswitch.
Nat.Chem.Biol., 10, 2014
|
|
1RO9
 
 | | CRYSTAL STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHODIESTERASE 4B2B COMPLEXED WITH 8-Br-AMP | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4B | | Authors: | Xu, R.X, Rocque, W.J, Lambert, M.H, Vanderwall, D.E, Nolte, R.T. | | Deposit date: | 2003-12-01 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram.
J.Mol.Biol., 337, 2004
|
|
5EZV
 
 | |
