1ULF
 
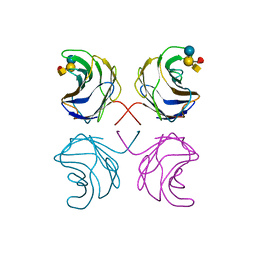 | | CGL2 in complex with Blood Group A tetrasaccharide | | Descriptor: | alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
6WI2
 
 | |
8YXR
 
 | |
5VYZ
 
 | | Crystal structure of Lactococcus lactis pyruvate carboxylase in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VZ0
 
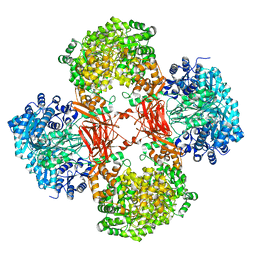 | | Crystal structure of Lactococcus lactis pyruvate carboxylase G746A mutant in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3E7K
 
 | |
6JY3
 
 | | Monomeric Form of Bovine Heart Cytochrome c Oxidase in the Fully Oxidized State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (2S,3S,4S,5S,6R)-2-(2-decoxyethoxy)-6-(hydroxymethyl)oxane-3,4,5-triol, ... | | Authors: | Shinzawa-Itoh, K, Muramoto, K. | | Deposit date: | 2019-04-26 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Monomeric structure of an active form of bovine cytochromecoxidase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ZKG
 
 | | Complex I with NADH, closed | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
4U8Y
 
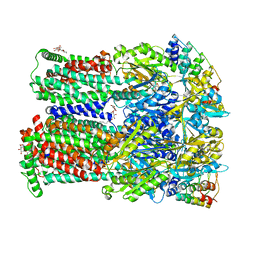 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pos, K.M. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB.
Elife, 3, 2014
|
|
8DNJ
 
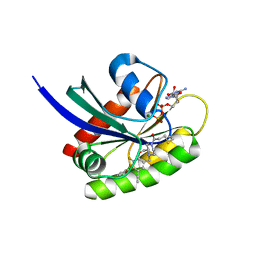 | | Crystal structure of human KRAS G12C covalently bound with AstraZeneca WO2020/178282A1 compound 76 | | Descriptor: | 1-[(5S,9P,12aR)-9-(2-chloro-6-hydroxyphenyl)-8-ethynyl-10-fluoro-3,4,12,12a-tetrahydro-6H-pyrazino[2,1-c][1,4]benzoxazepin-2(1H)-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7RAD
 
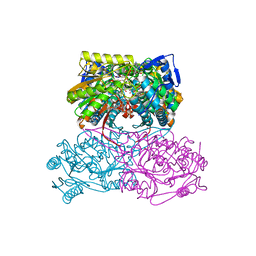 | | Crystal Structure Analysis of ALDH1B1 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-methoxyphenyl)-1-(4-phenylphenyl)-6,7,8,9-tetrahydro-5~{H}-imidazo[1,2-a][1,3]diazepine, Aldehyde dehydrogenase X, ... | | Authors: | Fernandez, D, Chen, J.K. | | Deposit date: | 2021-07-01 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors targeted to aldehyde dehydrogenase
Nat.Chem.Biol., 2022
|
|
6JST
 
 | |
5K7H
 
 | | Crystal structure of AibR in complex with the effector molecule isovaleryl coenzyme A | | Descriptor: | CHLORIDE ION, Isovaleryl-coenzyme A, NICKEL (II) ION, ... | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
6QC9
 
 | | Ovine respiratory complex I FRC open class 4 | | Descriptor: | Acyl carrier protein, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6K5O
 
 | | Development of Novel Lithocholic Acid Derivatives as Vitamin D Receptor Agonists | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3-methylsulfonyloxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Kagechika, H, Ito, N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel lithocholic acid derivatives as vitamin D receptor agonists.
Bioorg.Med.Chem., 27, 2019
|
|
6JWA
 
 | | Crystal structure of CK2a1 with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, Casein kinase II subunit alpha | | Authors: | Tsuyuguchi, M, Kinoshita, T. | | Deposit date: | 2019-04-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A promiscuous kinase inhibitor delineates the conspicuous structural features of protein kinase CK2a1.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6QCA
 
 | | Ovine respiratory complex I FRC open class 5 | | Descriptor: | Acyl carrier protein, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6QC3
 
 | | Ovine respiratory supercomplex I+III2 open class 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-26 | | Release date: | 2019-08-21 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6ZKI
 
 | | Complex I with NADH, open2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
7CMQ
 
 | |
6ZKH
 
 | | Complex I with NADH, open1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZKV
 
 | | Deactive complex I, open4 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
4U51
 
 | | Crystal structure of Narciclasine bound to the yeast 80S ribosome | | Descriptor: | (2S,3R,4S,4aR)-2,3,4,7-tetrahydroxy-3,4,4a,5-tetrahydro[1,3]dioxolo[4,5-j]phenanthridin-6(2H)-one, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
6K6T
 
 | | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - c-di-IMP bound form | | Descriptor: | 9-[(1R,6R,8R,9S,10R,15S,17R,18S)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-17-(6-oxidanylidene-3H-purin-9-yl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-8-yl]-3H-purin-6-one, CALCIUM ION, EAL domain protein | | Authors: | Yadav, M, Pal, K, Sen, U. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - c-di-IMP bound form
To Be Published
|
|
6KJR
 
 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | 4-[[(3~{E},5~{Z},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-14,16-bis(oxidanyl)-2-oxidanylidene-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-8-yl]oxy]-4-oxidanylidene-butanoic acid, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
