5QEB
 
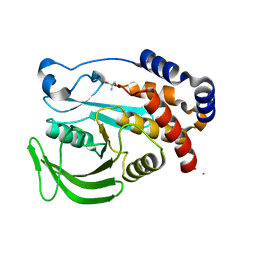 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000639a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloro-N~1~-(pyridin-4-yl)benzene-1,2-diamine, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5NSA
 
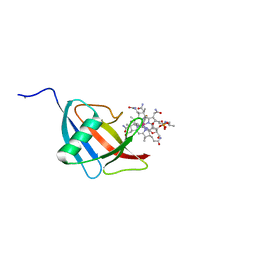 | | Beta domain of human transcobalamin bound to Co-beta-[2-(2,4-difluorophenyl)ethinyl]cobalamin | | Descriptor: | 1-ethynyl-2,4-difluorobenzene, CALCIUM ION, COBALAMIN, ... | | Authors: | Bloch, J.S, Ruetz, M, Krautler, B, Locher, K.P. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.273 Å) | | Cite: | Structure of the human transcobalamin beta domain in four distinct states.
PLoS ONE, 12, 2017
|
|
5QBF
 
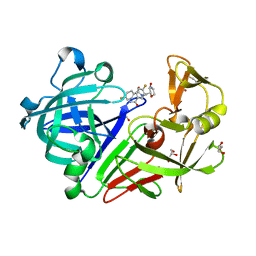 | | Crystal structure of Endothiapepsin-FRG203 complex | | Descriptor: | 2-amino-1,4-anhydro-2,5-dideoxy-5-[(4-fluorobenzene-1-carbonyl)amino]-D-arabinitol, ACETATE ION, Endothiapepsin, ... | | Authors: | Huschmann, F. | | Deposit date: | 2017-08-04 | | Release date: | 2020-04-22 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.268 Å) | | Cite: | Crystal structure of Endothiapepsin
To be published
|
|
5PZX
 
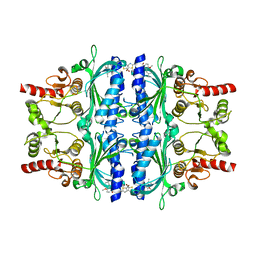 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-methoxy-3-(2-methylpropyl)phenyl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-4-methoxy-3-(2-methylpropyl)benzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-methoxy-3-(2-methylpropyl)phenyl]sulfonylurea
To be published
|
|
5QS5
 
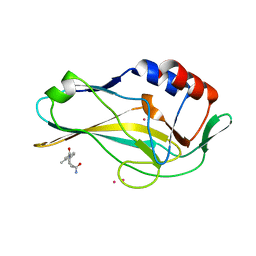 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury in complex with Z32400357 | | Descriptor: | 1-(3-methylbenzene-1-carbonyl)piperidine-4-carboxamide, CADMIUM ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Fernandez-Cid, A, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QTE
 
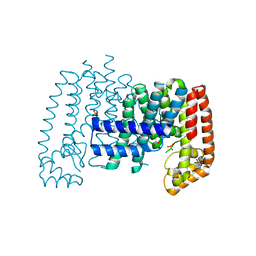 | | T. brucei FPPS in complex with CID 4563894 | | Descriptor: | (4S)-3-(4-chlorobenzene-1-carbonyl)-1,3-thiazolidine-4-carboxylic acid, Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Cornaciu, I, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-09 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | T. brucei FPPS in complex with CID 4563894
To Be Published
|
|
5QTK
 
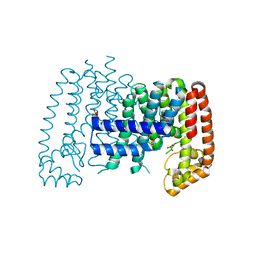 | | T. brucei FPPS in complex with CID 69539 | | Descriptor: | Farnesyl pyrophosphate synthase, N,N-diethyl-4-methylbenzene-1-sulfonamide, PHOSPHATE ION, ... | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Cornaciu, I, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-09 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | T. brucei FPPS in complex with CID 69539
To Be Published
|
|
5QPE
 
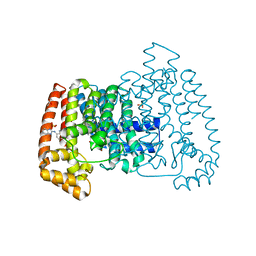 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000295a | | Descriptor: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5Q01
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(4,5-dichloro-2-methylpyrazol-3-yl)oxyphenyl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-4-[(3,4-dichloro-1-methyl-1H-pyrazol-5-yl)oxy]benzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(4,5-dichloro-2-methylpyrazol-3-yl)oxyphenyl]sulfonylurea
To be published
|
|
5QTG
 
 | | T. brucei FPPS in complex with CID 126782062 | | Descriptor: | 2-fluoro-N,3-dimethylbenzene-1-sulfonamide, Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Cornaciu, I, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-09 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | T. brucei FPPS in complex with CID 126782062
To Be Published
|
|
5PZZ
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-(3,4-dichlorophenyl)sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-3,4-dichlorobenzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-28 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-(3,4-dichlorophenyl)sulfonylurea
To be published
|
|
5Q0M
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-{[(3beta,5beta,14beta,17alpha)-3-hydroxy-24-oxocholan-24-yl]amino}benzene-1,3-dicarboxylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5PZR
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[3-[3-[(3-chlorophenyl)sulfonylcarbamoylamino]propoxy]propyl]urea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N,N'-{oxybis[(propane-3,1-diyl)carbamoyl]}bis(3-chlorobenzene-1-sulfonamide) | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[3-[3-[(3-chlorophenyl)sulfonylcarbamoylamino]propoxy]propyl]urea
To be published
|
|
5PZS
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[6-[(3-chlorophenyl)sulfonylcarbamoylamino]hexyl]urea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N,N'-(hexane-1,6-diyldicarbamoyl)bis(3-chlorobenzene-1-sulfonamide) | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[6-[(3-chlorophenyl)sulfonylcarbamoylamino]hexyl]urea
To be published
|
|
5Q06
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(2-methyl-1,3-thiazol-4-yl)phenyl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-4-(2-methyl-1,3-thiazol-4-yl)benzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(2-methyl-1,3-thiazol-4-yl)phenyl]sulfonylurea
To be published
|
|
5RB1
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001700a | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5R93
 
 | |
5RHX
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z1324080698 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-fluoro-5-methylbenzene-1-sulfonamide, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RZU
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z32400357 | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-methylbenzene-1-carbonyl)piperidine-4-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RXL
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1673618163 | | Descriptor: | 4-amino-N-(2-hydroxyethyl)-N-methylbenzene-1-sulfonamide, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5O39
 
 | | Human Brd2(BD2) mutant in complex with ME | | Descriptor: | (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, CHLORIDE ION, ... | | Authors: | Runcie, A.C, Chan, K.-H, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5O3F
 
 | | Human Brd2(BD2) mutant in complex with ET-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-butanamide, (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, ... | | Authors: | Runcie, A.C, Chan, K.-H, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5O68
 
 | | Crystal Structure of the Pseudomonas functional amyloid secretion protein FapF - R157A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5OD5
 
 | | Periplasmic binding protein CeuE complexed with a synthetic catalyst | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 4-(aminomethyl)-~{N}-(pyridin-2-ylmethyl)benzenesulfonamide, Azotochelin, ... | | Authors: | Duhme-Klair, A.K, Raines, D.J, Clarke, J.E, Blagova, E.V, Dodson, E.J, Wilson, K.S. | | Deposit date: | 2017-07-04 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox-switchable siderophore anchor enables reversible artificial metalloenzyme assembly
Nat Catal, 2018
|
|
5N8Y
 
 | | KaiCBA circadian clock backbone model based on a Cryo-EM density | | Descriptor: | Circadian clock protein KaiA, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Schuller, J.M, Snijder, J, Loessl, P, Heck, A.J.R, Foerster, F. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state.
Science, 355, 2017
|
|
