2WHL
 
 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
7E2U
 
 | | Synechocystis GUN4 in complex with phytochrome | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, ... | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
5V86
 
 | | Structure of DCN1 bound to NAcM-OPT | | Descriptor: | Lysozyme,DCN1-like protein 1, N-benzyl-N-(1-butylpiperidin-4-yl)-N'-(3,4-dichlorophenyl)urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
7EIR
 
 | | Crystal structure of chondroitin ABC lyase I in complex with chondroitin disaccharide 6S | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, Chondroitin sulfate ABC endolyase, GLYCEROL, ... | | Authors: | Takashima, M, Miyanaga, A, Eguchi, T. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate specificity of Chondroitinase ABC I based on analyses of biochemical reactions and crystal structures in complex with disaccharides.
Glycobiology, 31, 2021
|
|
5Z5O
 
 | | Structure of Pycnonodysostosis disease related I249T mutant of human cathepsin K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Biswas, S, Roy, S. | | Deposit date: | 2018-01-19 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Not all pycnodysostosis-related mutants of human cathepsin K are inactive - crystal structure and biochemical studies of an active mutant I249T.
FEBS J., 285, 2018
|
|
2WQ4
 
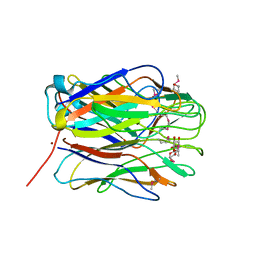 | | N-terminal domain of BC2L-C Lectin from Burkholderia cenocepacia | | Descriptor: | BROMIDE ION, LECTIN, methyl 1-seleno-alpha-L-fucopyranoside | | Authors: | Sulak, O, Cioci, G, Lahman, M, Delia, M, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2009-08-13 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Tnf-Like Trimeric Lectin Domain from Burkholderia Cenocepacia with Specificity for Fucosylated Human Histo-Blood Group Antigens
Structure, 18, 2010
|
|
5V84
 
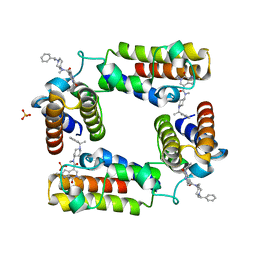 | | CECR2 in complex with Cpd6 (6-allyl-N,2-dimethyl-7-oxo-N-(1-(1-phenylethyl)piperidin-4-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4-carboxamide) | | Descriptor: | Cat eye syndrome critical region protein 2, N,2-dimethyl-7-oxo-N-{1-[(1S)-1-phenylethyl]piperidin-4-yl}-6-(prop-2-en-1-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4- carboxamide, SULFATE ION | | Authors: | Murray, J.M, Kiefer, J.R, Jayaran, H, Bellon, S, Boy, F. | | Deposit date: | 2017-03-21 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | GNE-886: A Potent and Selective Inhibitor of the Cat Eye Syndrome Chromosome Region Candidate 2 Bromodomain (CECR2).
ACS Med Chem Lett, 8, 2017
|
|
2CZL
 
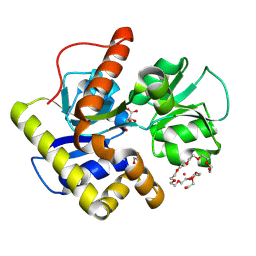 | | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8 (Cys11 modified with beta-mercaptoethanol) | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Arai, R, Nishino, A, Nagano, K, Kamo-Uchikubo, T, Nishimoto, M, Toyama, M, Terada, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8.
J.Struct.Biol., 168, 2009
|
|
7EFL
 
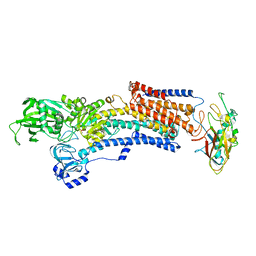 | | Crystal structure of the gastric proton pump K791S in (BYK)E2BeF state | | Descriptor: | (7R,8R,9S)-2,3-dimethyl-9-phenyl-7,8,9,10-tetrahydroimidazo[1,2-h][1,7]naphthyridine-7,8-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Abe, K, Yamamoto, K, Irie, K. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Gastric proton pump with two occluded K + engineered with sodium pump-mimetic mutations.
Nat Commun, 12, 2021
|
|
2WVV
 
 | | Crystal structure of an alpha-L-fucosidase GH29 from Bacteroides thetaiotaomicron | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-L-FUCOSIDASE, GLYCEROL, ... | | Authors: | Lammerts van Bueren, A, Ardevol, A, Fayers-Kerr, J, Luo, B, Zhang, Y, Sollogoub, M, Bleriot, Y, Rovira, C, Davies, G.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Analysis of the Reaction Coordinate of Alpha-L-Fucosidases: A Combined Structural and Quantum Mechanical Approach
J.Am.Chem.Soc., 132, 2010
|
|
5AKW
 
 | | Crystal structure of TNKS2 in complex with 2-(4-chlorophenyl)-1,2,3,4- tetrahydroquinazolin-4-one | | Descriptor: | (2S)-2-(4-chlorophenyl)-2,3-dihydro-1H-quinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Nkizinkiko, Y, Lehtio, L. | | Deposit date: | 2015-03-05 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Potent and Selective Nonplanar Tankyrase Inhibiting Nicotinamide Mimics.
Bioorg.Med.Chem., 23, 2015
|
|
7EFM
 
 | | Crystal structure of the gastric proton pump K791S/E820D/Y340N in (BYK)E2BeF state | | Descriptor: | (7R,8R,9S)-2,3-dimethyl-9-phenyl-7,8,9,10-tetrahydroimidazo[1,2-h][1,7]naphthyridine-7,8-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Abe, K, Yamamoto, K, Irie, K. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Gastric proton pump with two occluded K + engineered with sodium pump-mimetic mutations.
Nat Commun, 12, 2021
|
|
6N9Y
 
 | | Atomic structure of Non-Structural protein 1 of bluetongue virus | | Descriptor: | Non-structural protein 1 | | Authors: | Kerviel, A, Ge, P, Lai, M, Jih, J, Boyce, M, Zhang, X, Zhou, Z.H, Roy, P. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic structure of the translation regulatory protein NS1 of bluetongue virus.
Nat Microbiol, 4, 2019
|
|
4U71
 
 | |
5AE3
 
 | | Ether Lipid-Generating Enzyme AGPS in complex with antimycin A | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Piano, V, Benjamin, D.I, Valente, S, Nenci, S, Marrocco, B, Mai, A, Aliverti, A, Nomura, D.K, Mattevi, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Inhibitors for the Ether Lipid-Generating Enzyme Agps as Anti-Cancer Agents.
Acs Chem.Biol., 10, 2015
|
|
7EFN
 
 | | Crystal structure of the gastric proton pump K791S/E820D/Y340N/E936V in (BYK)E2BeF state | | Descriptor: | (7R,8R,9S)-2,3-dimethyl-9-phenyl-7,8,9,10-tetrahydroimidazo[1,2-h][1,7]naphthyridine-7,8-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Abe, K, Yamamoto, K, Irie, K. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Gastric proton pump with two occluded K + engineered with sodium pump-mimetic mutations.
Nat Commun, 12, 2021
|
|
5ZCG
 
 | | Crystal structure of OsPP2C50 S265L/I267V:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
4TZO
 
 | |
7E0A
 
 | | X-ray structure of human PPARgamma ligand binding domain-saroglitazar co-crystals obtained by co-crystallization | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Isoform 2 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Uchii, K, Machida, Y, Oyama, T, Ishii, I. | | Deposit date: | 2021-01-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Structural Basis for Anti-non-alcoholic Fatty Liver Disease and Diabetic Dyslipidemia Drug Saroglitazar as a PPAR alpha / gamma Dual Agonist.
Biol.Pharm.Bull., 44, 2021
|
|
7EBM
 
 | | W363A mutant of Chitin-specific solute binding protein from Vibrio harveyi in complex with chitobiose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
7EBI
 
 | | Chitin-specific solute binding protein from Vibrio harveyi co-crystalized with chitotetraose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-08 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
7GN5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-968bafd9-1 (Mpro-P2295) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(1-methoxycyclopropyl)methanesulfonyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5T8F
 
 | | p110delta/p85alpha with taselisib (GDC-0032) | | Descriptor: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Moertl, M, Steinbacher, S, Eigenbrot, C. | | Deposit date: | 2016-09-07 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
5ZHT
 
 | | Crystal structure of OsD14 in complex with covalently bound KK073 | | Descriptor: | (1H-1,2,3-triazol-1-yl){4-[4-(trifluoromethyl)phenyl]piperazin-1-yl}methanone, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
3I7R
 
 | | Dihydrodipicolinate synthase - K161R | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
